Comparing the taxonomic and functional profiles of gut microbiota from three pig breeds by metagenomic sequencing
- PMID: 36313418
- PMCID: PMC9614230
- DOI: 10.3389/fgene.2022.999535
Comparing the taxonomic and functional profiles of gut microbiota from three pig breeds by metagenomic sequencing
Abstract
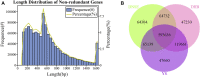
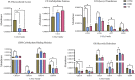
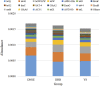
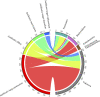
To investigate the difference of microbial communities among Diannan small-ear (DNSE), Dahe black (DHB) and Yorkshire (YS) pigs, we compared the microbial taxonomic and functional composition using a metagenomic approach. A total of 1,002,362 non-redundant microbial genes were identified, DHB and YS pigs had more similar genetic makeup compared with DNSE pigs. Bacteroidetes, Firmicutes and Spirochetes were the three most abundant phyla for all pig breeds, and DNSE pigs had a higher abundance of Prevotella genus than DHB and YS pigs. The functional profiles varied among the three pig breeds, DNSE pigs had more active carbohydrate metabolism and more abundant antibiotic resistance genes than the other two pig breeds. Moreover, we found that peptide and macrolide resistances genes in DNSE pigs were more abundant than that in DHB pigs (p < 0.05). This study will help to provide a theoretical basis for the development of native pig breeds in Yunnan Province, China.
Keywords: antibiotic resistance genes; carbohydrate metabolism; genetic makeup; metagenomic approach; microbial communities; pigs.
Copyright © 2022 Xu, Sun, Yi, Yang, Zhu, Huang, Pan, Li, Li, Zhao, Wei and Zhao.
Figures




Similar articles
-
Comparative Analyses of Antibiotic Resistance Genes in Jejunum Microbiota of Pigs in Different Areas.Front Cell Infect Microbiol. 2022 May 26;12:887428. doi: 10.3389/fcimb.2022.887428. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35719330 Free PMC article.
-
Comparison of the gut microbiota and metabolites between Diannan small ear pigs and Diqing Tibetan pigs.Front Microbiol. 2023 Jul 6;14:1197981. doi: 10.3389/fmicb.2023.1197981. eCollection 2023. Front Microbiol. 2023. PMID: 37485506 Free PMC article.
-
A comparison of dynamic distributions of intestinal microbiota between Large White and Chinese Shanxi Black pigs.Arch Microbiol. 2019 Apr;201(3):357-367. doi: 10.1007/s00203-019-01620-4. Epub 2019 Jan 23. Arch Microbiol. 2019. PMID: 30673796
-
Oropharyngeal, proximal colonic, and vaginal microbiomes of healthy Korean native black pig gilts.BMC Microbiol. 2023 Jan 5;23(1):3. doi: 10.1186/s12866-022-02743-3. BMC Microbiol. 2023. PMID: 36600197 Free PMC article.
-
Characterization of Metagenome-Assembled Genomes and Carbohydrate-Degrading Genes in the Gut Microbiota of Tibetan Pig.Front Microbiol. 2020 Dec 23;11:595066. doi: 10.3389/fmicb.2020.595066. eCollection 2020. Front Microbiol. 2020. PMID: 33424798 Free PMC article.
References
LinkOut - more resources
Full Text Sources

