Genome-wide identification of Aux/IAA gene family and their expression analysis in Prunus mume
- PMID: 36313426
- PMCID: PMC9597081
- DOI: 10.3389/fgene.2022.1013822
Genome-wide identification of Aux/IAA gene family and their expression analysis in Prunus mume
Erratum in
-
Corrigendum: Genome-wide identification of Aux/IAA gene family and their expression analysis in Prunus mume.Front Genet. 2023 May 9;14:1208488. doi: 10.3389/fgene.2023.1208488. eCollection 2023. Front Genet. 2023. PMID: 37229203 Free PMC article.
Abstract
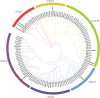
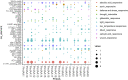
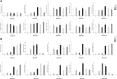
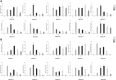
AUXIN/INDOLE ACETIC ACIDs (Aux/IAAs), an early auxin-responsive gene family, is important for plant growth and development. To fully comprehend the character of Aux/IAA genes in woody plants, we identified 19 PmIAA genes in Prunus mume and dissected their protein domains, phylogenetic relationship, gene structure, promoter, and expression patterns during floral bud flushing, auxin response, and abiotic stress response. The study showed that PmIAA proteins shared conserved Aux/IAA domain, but differed in protein motif composition. 19 PmIAA genes were divided into six groups (Groups Ⅰ to Ⅵ) based on phylogenetic analysis. The gene duplication analysis showed that segmental and dispersed duplication greatly influenced the expansion of PmIAA genes. Moreover, we identified and classified the cis-elements of PmIAA gene promoters and detected elements that are related to phytohormone responses and abiotic stress responses. With expression pattern analysis, we observed the auxin-responsive expression of PmIAA5, PmIAA17, and PmIAA18 in flower bud, stem, and leaf tissues. PmIAA5, PmIAA13, PmIAA14, and PmIAA18 were possibly involved in abiotic stress responses in P. mume. In general, these results laid the theoretical foundation for elaborating the functions of Aux/IAA genes in perennial woody plant development.
Keywords: Aux/IAA gene family; Prunus mume; auxin-responsive genes; evolutionary analysis; expression pattern analysis.
Copyright © 2022 Cheng, Zhang, Cheng, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Genome-wide identification and expression pattern analysis of the Aux/IAA (auxin/indole-3-acetic acid) gene family in alfalfa (Medicago sativa) and the potential functions under drought stress.BMC Genomics. 2024 Apr 18;25(1):382. doi: 10.1186/s12864-024-10313-2. BMC Genomics. 2024. PMID: 38637768 Free PMC article.
-
Bioinformatics Study of Aux/IAA Family Genes and Their Expression in Response to Different Hormones Treatments during Japanese Apricot Fruit Development and Ripening.Plants (Basel). 2022 Jul 22;11(15):1898. doi: 10.3390/plants11151898. Plants (Basel). 2022. PMID: 35893602 Free PMC article.
-
Genome-wide identification and salt stress-responsive expression profiling of Aux/IAA gene family in Asparagus officinalis.BMC Plant Biol. 2025 Jun 4;25(1):759. doi: 10.1186/s12870-025-06780-8. BMC Plant Biol. 2025. PMID: 40468198 Free PMC article.
-
Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function.Int J Mol Sci. 2018 Jan 16;19(1):259. doi: 10.3390/ijms19010259. Int J Mol Sci. 2018. PMID: 29337875 Free PMC article. Review.
-
Advances in structure and function of auxin response factor in plants.J Integr Plant Biol. 2023 Mar;65(3):617-632. doi: 10.1111/jipb.13392. Epub 2022 Dec 31. J Integr Plant Biol. 2023. PMID: 36263892 Review.
Cited by
-
Transcriptomic analysis of wrinkled leaf development of Tai-cai (Brassica rapa var. tai-tsai) and its synthetic allotetraploid via RNA and miRNA sequencing.Plant Mol Biol. 2025 May 6;115(3):66. doi: 10.1007/s11103-025-01592-8. Plant Mol Biol. 2025. PMID: 40327156
-
Transcriptome profiling reveals key regulatory factors and metabolic pathways associated with curd formation and development in broccoli.Front Plant Sci. 2024 Jul 12;15:1418319. doi: 10.3389/fpls.2024.1418319. eCollection 2024. Front Plant Sci. 2024. PMID: 39070909 Free PMC article.
-
Genome-wide identification and expression pattern analysis of the Aux/IAA (auxin/indole-3-acetic acid) gene family in alfalfa (Medicago sativa) and the potential functions under drought stress.BMC Genomics. 2024 Apr 18;25(1):382. doi: 10.1186/s12864-024-10313-2. BMC Genomics. 2024. PMID: 38637768 Free PMC article.
-
Genome-wide identification of Aux/IAA gene family members in grape and functional analysis of VaIAA3 in response to cold stress.Plant Cell Rep. 2024 Oct 17;43(11):265. doi: 10.1007/s00299-024-03353-1. Plant Cell Rep. 2024. PMID: 39417869
-
Whole-Genome Identification of Regulatory Function of CDPK Gene Families in Cold Stress Response for Prunus mume and Prunus mume var. Tortuosa.Plants (Basel). 2023 Jul 4;12(13):2548. doi: 10.3390/plants12132548. Plants (Basel). 2023. PMID: 37447109 Free PMC article.
References
-
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. - PubMed
LinkOut - more resources
Full Text Sources

