Insight into selective breeding for robustness based on field survival records: New genetic evaluation of survival traits in pacific white shrimp (Penaeus vannamei) breeding line
- PMID: 36313448
- PMCID: PMC9608658
- DOI: 10.3389/fgene.2022.1018568
Insight into selective breeding for robustness based on field survival records: New genetic evaluation of survival traits in pacific white shrimp (Penaeus vannamei) breeding line
Abstract
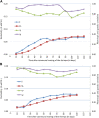
Survival can be considered a relatively 'old' trait in animal breeding, yet commonly neglected in aquaculture breeding because of the simple binary records and generally low heritability estimates. Developing routine genetic evaluation systems for survival traits however, will be important for breeding robust strains based on valuable field survival data. In the current study, linear multivariate animal model (LMA) was used for the genetic analysis of survival records from 2-year classes (BL2019 and BL2020) of pacific white shrimp (Penaeus vannamei) breeding lines with data collection of 52, 248 individuals from 481 fullsib families. During grow-out test period, 10 days intervals of survival data were considered as separate traits. Two survival definitions, binary survivability (S) and continuous survival in days (SL), were used for the genetic analysis of survival records to investigate; 1) whether adding more survival time information could improve estimation of genetic parameters; 2) the trajectory of survival heritability across time, and 3) patterns of genetic correlations of survival traits across time. Levels of heritability estimates for both S and SL were low (0.005-0.076), while heritability for survival day number was found to be similar with that of binary records at each observation time and were highly genetically correlated (r g > 0.8). Heritability estimates of body weight (BW) for BL2019 and BL2020 were 0.486 and 0.373, respectively. Trajectories of survival heritability showed a gradual increase across the grow-out test period but slowed or reached a plateau during the later grow-out test period. Genetic correlations among survival traits in the grow-out tests were moderate to high, and the closer the times were between estimates, the higher were their genetic correlations. In contrast, genetic correlations between both survival traits and body weight were low but positive. Here we provide the first report on the trajectory of heritability estimates for survival traits across grow-out stage in aquaculture. Results will be useful for developing robust improved pacific white shrimp culture strains in selective breeding programs based on field survival data.
Keywords: Penaeus vannamei; genetic evaluation; genetic parameters; heritability; selective breeding; survival traits.
Copyright © 2022 Ren, Mather, Tang and Hurwood.
Conflict of interest statement
BT is employed by the Company Beijing Shuishiji Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bangera R., Ødegård J., Mikkelsen H., Nielsen H. M., Seppola M., Puvanendran V., et al. (2014). Genetic analysis of francisellosis field outbreak in Atlantic cod (Gadus morhua L.) using an ordinal threshold model. Aquaculture 420, S50–S56. 10.1016/j.aquaculture.2013.08.029 - DOI
-
- Barría A., Christensen K. A., Yoshida G., Jedlicki A., Leong J. S., Rondeau E. B., et al. (2019). Whole genome linkage disequilibrium and effective population size in a coho salmon (Oncorhynchus kisutch) breeding population using a high-density SNP array. Front. Genet. 10, 498. 10.3389/fgene.2019.00498 - DOI - PMC - PubMed
-
- Barría A., Trinh T. Q., Mahmuddin M., Benzie J. A., Chadag V. M., Houston R. D. (2020). Genetic parameters for resistance to Tilapia lake virus (TiLV) in nile tilapia (Oreochromis niloticus). Aquaculture 522, 735126. 10.1016/j.aquaculture.2020.735126 - DOI
LinkOut - more resources
Full Text Sources

