Nramp: Deprive and conquer?
- PMID: 36313567
- PMCID: PMC9606685
- DOI: 10.3389/fcell.2022.988866
Nramp: Deprive and conquer?
Abstract
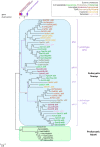
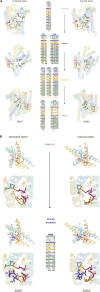
Solute carriers 11 (Slc11) evolved from bacterial permease (MntH) to eukaryotic antibacterial defense (Nramp) while continuously mediating proton (H+)-dependent manganese (Mn2+) import. Also, Nramp horizontal gene transfer (HGT) toward bacteria led to mntH polyphyly. Prior demonstration that evolutionary rate-shifts distinguishing Slc11 from outgroup carriers dictate catalytic specificity suggested that resolving Slc11 family tree may provide a function-aware phylogenetic framework. Hence, MntH C (MC) subgroups resulted from HGTs of prototype Nramp (pNs) parologs while archetype Nramp (aNs) correlated with phagocytosis. PHI-Blast based taxonomic profiling confirmed MntH B phylogroup is confined to anaerobic bacteria vs. MntH A (MA)'s broad distribution; suggested niche-related spread of MC subgroups; established that MA-variant MH, which carries 'eukaryotic signature' marks, predominates in archaea. Slc11 phylogeny shows MH is sister to Nramp. Site-specific analysis of Slc11 charge network known to interact with the protonmotive force demonstrates sequential rate-shifts that recapitulate Slc11 evolution. 3D mapping of similarly coevolved sites across Slc11 hydrophobic core revealed successive targeting of discrete areas. The data imply that pN HGT could advantage recipient bacteria for H+-dependent Mn2+ acquisition and Alphafold 3D models suggest conformational divergence among MC subgroups. It is proposed that Slc11 originated as a bacterial stress resistance function allowing Mn2+-dependent persistence in conditions adverse for growth, and that archaeal MH could contribute to eukaryogenesis as a Mn2+ sequestering defense perhaps favoring intracellular growth-competent bacteria.
Keywords: MntH (proton-dependent Mn transporter); Nramp (natural resistance-associated macrophage protein); divalent metal-ions; evolutionary rate-shift; horizontal gene transfer; molecular phylogeny; nutritional immunity; protonmotive force.
Copyright © 2022 Cellier.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

