CasPlay provides a gRNA-barcoded CRISPR-based display platform for antibody repertoire profiling
- PMID: 36313802
- PMCID: PMC9606310
- DOI: 10.1016/j.crmeth.2022.100318
CasPlay provides a gRNA-barcoded CRISPR-based display platform for antibody repertoire profiling
Abstract
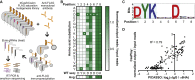
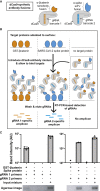
Protein display technologies link proteins to distinct nucleic acid sequences (barcodes), enabling multiplexed protein assays via DNA sequencing. Here, we develop Cas9 display (CasPlay) to interrogate customized peptide libraries fused to catalytically inactive Cas9 (dCas9) by sequencing the guide RNA (gRNA) barcodes associated with each peptide. We first confirm the ability of CasPlay to characterize antibody epitopes by recovering a known binding motif for a monoclonal anti-FLAG antibody. We then use a CasPlay library tiling the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteome to evaluate vaccine-induced antibody reactivities. Using a peptide library representing the human virome, we demonstrate the ability of CasPlay to identify epitopes across many viruses from microliters of patient serum. Our results suggest that CasPlay is a viable strategy for customized protein interaction studies from highly complex libraries and could provide an alternative to phage display technologies.
Keywords: CRISPR; Cas9; antibody binding; biotechnology; peptide display; protein display.
© 2022 The Author(s).
Conflict of interest statement
K.W.B. and S.J.E. have filed a patent application with the US Patent and Trademark Office (US patent application no. 63/220,399) related to this work. S.J.E. is a founder of TScan Therapeutics, Maze Therapeutics, ImmuneID, and Mirimus. S.J.E. is a scientific advisory board member for Homology Medicines, TScan Therapeutics, ImmuneID, and Maze Therapeutics.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

