DNA extraction bias is more pronounced for microbial eukaryotes than for prokaryotes
- PMID: 36314757
- PMCID: PMC9524606
- DOI: 10.1002/mbo3.1323
DNA extraction bias is more pronounced for microbial eukaryotes than for prokaryotes
Abstract
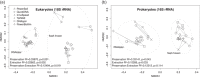
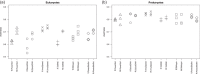
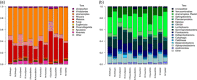
DNA extraction and preservation bias is a recurring topic in DNA sequencing-based microbial ecology. The different methodologies can lead to distinct outcomes, which has been demonstrated especially in studies investigating prokaryotic community composition. Eukaryotic microbes are ubiquitous, diverse, and increasingly a subject of investigation in addition to bacteria and archaea. However, little is known about how the choice of DNA preservation and extraction methodology impacts perceived eukaryotic community composition. In this study, we compared the effect of two DNA preservation methods and six DNA extraction methods on the community profiles of both eukaryotes and prokaryotes in phototrophic biofilms on seagrass (Zostera marina) leaves from the Baltic Sea. We found that, whereas both DNA preservation and extraction method caused significant bias in perceived community composition for both eukaryotes and prokaryotes, extraction bias was more pronounced for eukaryotes than for prokaryotes. In particular, soft-bodied and hard-shelled eukaryotes like nematodes and diatoms, respectively, were differentially abundant depending on the extraction method. We conclude that careful consideration of DNA preservation and extraction methodology is crucial to achieving representative community profiles of eukaryotes in marine biofilms and likely all other habitats containing diverse eukaryotic microbial communities.
Keywords: 18S rRNA; DNA extraction bias; DNA preservation; biofilm; microbial communities; seagrass microbiome.
© 2022 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures




References
-
- Burgunter‐Delamare, B. , Tanguy, G. , Legeay, E. , Boyen, C. , & Dittami, S. M. (2022). Effects of sampling and storage procedures on 16S rDNA amplicon sequencing results of kelp microbiomes. Marine Genomics, 63, 100944. - PubMed
-
- Delmont, T. O. , Gaia, M. , Hinsinger, D. D. , Frémont, P. , Vanni, C. , Fernandez‐Guerra, A , Eren, A. M. , Kourlaiev, A. , d'Agata, L. , Clayssen, Q. , Villar, E. , Labadie, K. , Cruaud, C. , Poulain, J. , Da Silva, C. , Wessner, M. , Noel, B. , Aury, J. M. , de Vargas, C. , … Speich, S. (2022). Functional repertoire convergence of distantly related eukaryotic plankton lineages revealed by genome‐resolved metagenomics. Cell Genomics, 2, 100123. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

