eIF4A3 Promotes RNA Viruses' Replication by Inhibiting Innate Immune Responses
- PMID: 36314820
- PMCID: PMC9683021
- DOI: 10.1128/jvi.01513-22
eIF4A3 Promotes RNA Viruses' Replication by Inhibiting Innate Immune Responses
Abstract
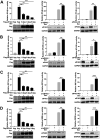
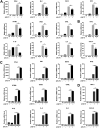
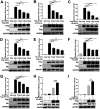
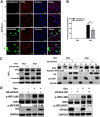
Viral infection activates the type I interferons (IFNs) and cellular antiviral responses. Eukaryotic initiation factor 4A-III (eIF4A3) has been shown to promote influenza A virus (IAV) replication by promoting viral mRNA splicing and spliced mRNA nuclear export. Here, we identified eIF4A3 as a negative regulator of virus-triggered type I IFN induction. Our study found that eIF4A3 promoted multiple RNA viruses' replication by binding to IFN regulatory factor 3 (IRF3) and impaired the interaction between tank-binding kinase 1 (TBK1) and IRF3, leading to attenuation of the phosphorylation of IRF3 by TBK1, the formation of IRF3 dimer, and the nuclear translocation of IRF3. This impaired its biological functions in the nucleus, which blocked IRF3 binding to interferon-stimulated response element (ISRE) and the interaction of IRF3 and CBP/p300, resulting in inhibiting the transcription of IFN-β and downstream IFN-stimulated genes (ISGs), thereby impairing innate antiviral immune responses against RNA viruses. These findings reveal a previously unknown function of eIF4A3 in host innate immunity and establish a mechanistic link between eIF4A3 and IRF3 activation that expands potential therapeutic strategies for viral infectious diseases. IMPORTANCE Production of type I IFN is pivotal for the cellular antiviral immunity. Virus infection leads to the activation of transcription factor IRF3 and subsequent production of type I IFN to eliminate viral infection. Thus, the regulation of IRF3 activity is an important way to affect type I IFN production. IRF3 activation requires phosphorylation, dimerization, and nuclear translocation. Here, we first reported that eIF4A3, a member of DEAD box family, served as a negative regulator of antiviral innate immune responses by inhibiting IRF3 activation. Mechanistically, eIF4A3 binds to IRF3 to impair the recruitment of IRF3 by TBK1, which is independent of eIF4A3 ATP binding, ATPase, and RNA helicase activities. Our study delineates a common mechanism of eIF4A3 promoting replication of different RNA viruses and provides important insights into the negative regulation of host antiviral innate immune responses against virus infections.
Keywords: IFN-β; IRF3; eIF4A3; innate immune responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Zinc-finger CCHC-type containing protein 8 promotes RNA virus replication by suppressing the type-I interferon responses.J Virol. 2024 Sep 17;98(9):e0079624. doi: 10.1128/jvi.00796-24. Epub 2024 Aug 8. J Virol. 2024. PMID: 39115433 Free PMC article.
-
The Molecular Mechanism of Herpes Simplex Virus 1 UL31 in Antagonizing the Activity of IFN-β.Microbiol Spectr. 2022 Feb 23;10(1):e0188321. doi: 10.1128/spectrum.01883-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196784 Free PMC article.
-
LYAR Suppresses Beta Interferon Induction by Targeting Phosphorylated Interferon Regulatory Factor 3.J Virol. 2019 Oct 15;93(21):e00769-19. doi: 10.1128/JVI.00769-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31413131 Free PMC article.
-
TANK-binding kinase 1 as a novel therapeutic target for viral diseases.Expert Opin Ther Targets. 2019 May;23(5):437-446. doi: 10.1080/14728222.2019.1601702. Epub 2019 Apr 10. Expert Opin Ther Targets. 2019. PMID: 30932713 Review.
-
Regulation of IRF3 activation in human antiviral signaling pathways.Biochem Pharmacol. 2022 Jun;200:115026. doi: 10.1016/j.bcp.2022.115026. Epub 2022 Mar 31. Biochem Pharmacol. 2022. PMID: 35367198 Review.
Cited by
-
Zinc-finger CCHC-type containing protein 8 promotes RNA virus replication by suppressing the type-I interferon responses.J Virol. 2024 Sep 17;98(9):e0079624. doi: 10.1128/jvi.00796-24. Epub 2024 Aug 8. J Virol. 2024. PMID: 39115433 Free PMC article.
-
Cervical mucus in linked human Cervix and Vagina Chips modulates vaginal dysbiosis.NPJ Womens Health. 2025;3(1):5. doi: 10.1038/s44294-025-00054-2. Epub 2025 Jan 29. NPJ Womens Health. 2025. PMID: 39896100 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

