Investigating Interactions Between Histone Modifying Enzymes and Transcription Factors in vivo by Fluorescence Resonance Energy Transfer
- PMID: 36314833
- PMCID: PMC9629860
- DOI: 10.3791/64656
Investigating Interactions Between Histone Modifying Enzymes and Transcription Factors in vivo by Fluorescence Resonance Energy Transfer
Abstract
Epigenetic regulation of gene expression is commonly affected by histone modifying enzymes (HMEs) that generate heterochromatic or euchromatic histone marks for transcriptional repression or activation, respectively. HMEs are recruited to their target chromatin by transcription factors (TFs). Thus, detecting and characterizing direct interactions between HMEs and TFs are critical for understanding their function and specificity better. These studies would be more biologically relevant if performed in vivo within living tissues. Here, a protocol is described for visualizing interactions in plant leaves between a plant histone deubiquitinase and a plant transcription factor using fluorescence resonance energy transfer (FRET), which allows the detection of complexes between protein molecules that are within <10 nm from each other. Two variations of the FRET technique are presented: SE-FRET (sensitized emission) and AB-FRET (acceptor bleaching), in which the energy is transferred non-radiatively from the donor to the acceptor or emitted radiatively by the donor upon photobleaching of the acceptor. Both SE-FRET and AB-FRET approaches can be adapted easily to discover other interactions between other proteins in planta.
Conflict of interest statement
DISCLOSURES:
No conflicts of interest were declared.
Figures

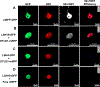
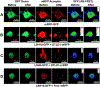
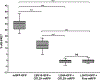
Similar articles
-
Analysis of photobleaching in single-molecule multicolor excitation and Förster resonance energy transfer measurements.J Phys Chem A. 2006 Mar 9;110(9):2979-95. doi: 10.1021/jp054581w. J Phys Chem A. 2006. PMID: 16509620
-
Photobleaching and Sensitized Emission-Based Methods for the Detection of Förster Resonance Energy Transfer.Methods Mol Biol. 2019;2040:235-274. doi: 10.1007/978-1-4939-9686-5_12. Methods Mol Biol. 2019. PMID: 31432483
-
Fluorescence resonance energy transfer of GFP and YFP by spectral imaging and quantitative acceptor photobleaching.J Microsc. 2008 Jul;231(Pt 1):97-104. doi: 10.1111/j.1365-2818.2008.02020.x. J Microsc. 2008. PMID: 18638193
-
Time-Resolved Fluorescence Resonance Energy Transfer [TR-FRET] Assays for Biochemical Processes.Curr Pharm Biotechnol. 2016;17(14):1222-1230. doi: 10.2174/1389201017666160809164527. Curr Pharm Biotechnol. 2016. PMID: 27604358 Review.
-
Protein localization in living cells and tissues using FRET and FLIM.Differentiation. 2003 Dec;71(9-10):528-41. doi: 10.1111/j.1432-0436.2003.07109007.x. Differentiation. 2003. PMID: 14686950 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
