Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection
- PMID: 36315049
- PMCID: PMC9623343
- DOI: 10.1084/jem.20220679
Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection
Abstract
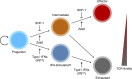
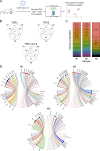
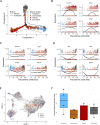
Although recent evidence demonstrates heterogeneity among CD8+ T cells during chronic infection, developmental relationships and mechanisms underlying their fate decisions remain incompletely understood. Using single-cell RNA and TCR sequencing, we traced the clonal expansion and differentiation of CD8+ T cells during chronic LCMV infection. We identified immense clonal and phenotypic diversity, including a subset termed intermediate cells. Trajectory analyses and infection models showed intermediate cells arise from progenitor cells before bifurcating into terminal effector and exhausted subsets. Genetic ablation experiments identified that type I IFN drives exhaustion through an IRF7-dependent mechanism, possibly through an IFN-stimulated subset bridging progenitor and exhausted cells. Conversely, Zeb2 was critical for generating effector cells. Intriguingly, some T cell clones exhibited lineage bias. Mechanistically, we identified that TCR avidity correlates with an exhausted fate, whereas SHP-1 selectively restricts low-avidity effector cell accumulation. Thus, our work elucidates novel mechanisms underlying CD8+ T cell fate determination during persistent infection and suggests two potential pathways leading to exhaustion.
© 2022 Kasmani et al.
Conflict of interest statement
Disclosures: S.M. Kaech reported personal fees from EvolveImmune Therapeutics, Affini-T Therapeutics, Arvinas, Pfizer, and Barer Institute outside the submitted work. No other disclosures were reported.
Figures












References
-
- Ahmadzadeh, M., Johnson L.A., Heemskerk B., Wunderlich J.R., Dudley M.E., White D.E., and Rosenberg S.A.. 2009. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 114:1537–1544. 10.1182/blood-2008-12-195792 - DOI - PMC - PubMed
-
- Alford, R.F., Leaver-Fay A., Jeliazkov J.R., O’Meara M.J., DiMaio F.P., Park H., Shapovalov M.V., Renfrew P.D., Mulligan V.K., Kappel K., et al. . 2017. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theor. Comput. 13:3031–3048. 10.1021/acs.jctc.7b00125 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI125741/AI/NIAID NIH HHS/United States
- R01 CA206483/CA/NCI NIH HHS/United States
- F30 CA246920/CA/NCI NIH HHS/United States
- R00 AI153537/AI/NIAID NIH HHS/United States
- R01 AI148403/AI/NIAID NIH HHS/United States
- T32-GM080202/GM/NIGMS NIH HHS/United States
- T32 GM080202/GM/NIGMS NIH HHS/United States
- AI125741/NH/NIH HHS/United States
- R01 AI066232/AI/NIAID NIH HHS/United States
- R37 AI066232/AI/NIAID NIH HHS/United States
- F30 DK127526/DK/NIDDK NIH HHS/United States
- R01 AI165578/AI/NIAID NIH HHS/United States
- K99 AI153537/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

