Tra1 controls the transcriptional landscape of the aging cell
- PMID: 36315064
- PMCID: PMC9836359
- DOI: 10.1093/g3journal/jkac287
Tra1 controls the transcriptional landscape of the aging cell
Abstract
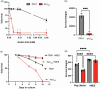
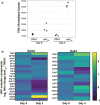
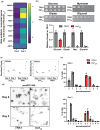
Gene expression undergoes considerable changes during the aging process. The mechanisms regulating the transcriptional response to cellular aging remain poorly understood. Here, we employ the budding yeast Saccharomyces cerevisiae to better understand how organisms adapt their transcriptome to promote longevity. Chronological lifespan assays in yeast measure the survival of nondividing cells at stationary phase over time, providing insights into the aging process of postmitotic cells. Tra1 is an essential component of both the yeast Spt-Ada-Gcn5 acetyltransferase/Spt-Ada-Gcn5 acetyltransferase-like and nucleosome acetyltransferase of H4 complexes, where it recruits these complexes to acetylate histones at targeted promoters. Importantly, Tra1 regulates the transcriptional response to multiple stresses. To evaluate the role of Tra1 in chronological aging, we took advantage of a previously characterized mutant allele that carries mutations in the TRA1 PI3K domain (tra1Q3). We found that loss of functions associated with tra1Q3 sensitizes cells to growth media acidification and shortens lifespan. Transcriptional profiling reveals that genes differentially regulated by Tra1 during the aging process are enriched for components of the response to stress. Notably, expression of catalases (CTA1, CTT1) involved in hydrogen peroxide detoxification decreases in chronologically aged tra1Q3 cells. Consequently, they display increased sensitivity to oxidative stress. tra1Q3 cells are unable to grow on glycerol indicating a defect in mitochondria function. Aged tra1Q3 cells also display reduced expression of peroxisomal genes, exhibit decreased numbers of peroxisomes, and cannot grow on media containing oleate. Thus, Tra1 emerges as an important regulator of longevity in yeast via multiple mechanisms.
Keywords: SAGA complex; Tra1; chronological aging; peroxisomes; yeast.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
None declared.
Figures




Similar articles
-
Evolutionary diversity of the control of the azole response by Tra1 across yeast species.G3 (Bethesda). 2024 Feb 7;14(2):jkad250. doi: 10.1093/g3journal/jkad250. G3 (Bethesda). 2024. PMID: 37889998 Free PMC article.
-
The SAGA and NuA4 component Tra1 regulates Candida albicans drug resistance and pathogenesis.Genetics. 2021 Oct 2;219(2):iyab131. doi: 10.1093/genetics/iyab131. Genetics. 2021. PMID: 34849885 Free PMC article.
-
Sfp1 links TORC1 and cell growth regulation to the yeast SAGA-complex component Tra1 in response to polyQ proteotoxicity.Traffic. 2019 Apr;20(4):267-283. doi: 10.1111/tra.12637. Traffic. 2019. PMID: 30740854
-
The Gcn5 complexes in Drosophila as a model for metazoa.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194610. doi: 10.1016/j.bbagrm.2020.194610. Epub 2020 Jul 28. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32735945 Review.
-
The SAGA continues: The rise of cis- and trans-histone crosstalk pathways.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194600. doi: 10.1016/j.bbagrm.2020.194600. Epub 2020 Jul 6. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32645359 Free PMC article. Review.
Cited by
-
Evolutionary diversity of the control of the azole response by Tra1 across yeast species.G3 (Bethesda). 2024 Feb 7;14(2):jkad250. doi: 10.1093/g3journal/jkad250. G3 (Bethesda). 2024. PMID: 37889998 Free PMC article.
-
Csn5 Depletion Reverses Mitochondrial Defects in GCN5-Null Saccharomyces cerevisiae.Int J Mol Sci. 2025 Jul 18;26(14):6916. doi: 10.3390/ijms26146916. Int J Mol Sci. 2025. PMID: 40725162 Free PMC article.
-
Live while the DNA lasts. The role of autophagy in DNA loss and survival of diploid yeast cells during chronological aging.Aging (Albany NY). 2023 Oct 9;15(19):9965-9983. doi: 10.18632/aging.205102. Epub 2023 Oct 9. Aging (Albany NY). 2023. PMID: 37815879 Free PMC article.
References
-
- Agarwal S, Sharma S, Agrawal V, Roy N.. Caloric restriction augments ROS defense in S. cerevisiae, by a Sir2p independent mechanism. Free Radic Res. 2005;39(1):55–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
