A family of methyl esterases converts methyl salicylate to salicylic acid in ripening tomato fruit
- PMID: 36315067
- PMCID: PMC9806648
- DOI: 10.1093/plphys/kiac509
A family of methyl esterases converts methyl salicylate to salicylic acid in ripening tomato fruit
Abstract
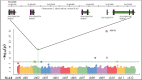
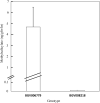
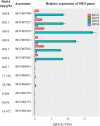
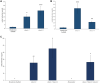
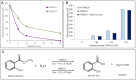
Methyl salicylate imparts a potent flavor and aroma described as medicinal and wintergreen that is undesirable in tomato (Solanum lycopersicum) fruit. Plants control the quantities of methyl salicylate through a variety of biosynthetic pathways, including the methylation of salicylic acid to form methyl salicylate and subsequent glycosylation to prevent methyl salicylate emission. Here, we identified a subclade of tomato methyl esterases, SALICYLIC ACID METHYL ESTERASE1-4, responsible for demethylation of methyl salicylate to form salicylic acid in fruits. This family was identified by proximity to a highly significant methyl salicylate genome-wide association study locus on chromosome 2. Genetic mapping studies in a biparental population confirmed a major methyl salicylate locus on chromosome 2. Fruits from SlMES1 knockout lines emitted significantly (P < 0,05, t test) higher amounts of methyl salicylate than wild-type fruits. Double and triple mutants of SlMES2, SlMES3, and SlMES4 emitted even more methyl salicylate than SlMES1 single knockouts-but not at statistically distinguishable levels-compared to the single mutant. Heterologously expressed SlMES1 and SlMES3 acted on methyl salicylate in vitro, with SlMES1 having a higher affinity for methyl salicylate than SlMES3. The SlMES locus has undergone major rearrangement, as demonstrated by genome structure analysis in the parents of the biparental population. Analysis of accessions that produce high or low levels of methyl salicylate showed that SlMES1 and SlMES3 genes expressed the highest in the low methyl salicylate lines. None of the MES genes were appreciably expressed in the high methyl salicylate-producing lines. We concluded that the SlMES gene family encodes tomato methyl esterases that convert methyl salicylate to salicylic acid in ripe tomato fruit. Their ability to decrease methyl salicylate levels by conversion to salicylic acid is an attractive breeding target to lower the level of a negative contributor to flavor.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Similar articles
-
Structural variation underlies functional diversity at methyl salicylate loci in tomato.PLoS Genet. 2023 May 4;19(5):e1010751. doi: 10.1371/journal.pgen.1010751. eCollection 2023 May. PLoS Genet. 2023. PMID: 37141297 Free PMC article.
-
SlMES1 modulates methyl salicylate to influence fruit volatile profiles in tomato.Plant Physiol Biochem. 2025 Apr;221:109561. doi: 10.1016/j.plaphy.2025.109561. Epub 2025 Feb 3. Plant Physiol Biochem. 2025. PMID: 39933427
-
Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate.Plant J. 2010 Apr 1;62(1):113-23. doi: 10.1111/j.1365-313X.2010.04128.x. Epub 2010 Jan 7. Plant J. 2010. PMID: 20070566
-
Proteomics of Reproductive Development, Fruit Ripening, and Stress Responses in Tomato.J Agric Food Chem. 2023 Jan 11;71(1):65-95. doi: 10.1021/acs.jafc.2c06564. Epub 2022 Dec 30. J Agric Food Chem. 2023. PMID: 36584279 Review.
-
Skin colour, carotenogenesis and chlorophyll degradation mutant alleles: genetic orchestration behind the fruit colour variation in tomato.Plant Cell Rep. 2021 May;40(5):767-782. doi: 10.1007/s00299-020-02650-9. Epub 2021 Jan 3. Plant Cell Rep. 2021. PMID: 33388894 Review.
Cited by
-
Recent Advances in Studying the Regulation of Fruit Ripening in Tomato Using Genetic Engineering Approaches.Int J Mol Sci. 2024 Jan 7;25(2):760. doi: 10.3390/ijms25020760. Int J Mol Sci. 2024. PMID: 38255834 Free PMC article. Review.
-
Structural variation underlies functional diversity at methyl salicylate loci in tomato.PLoS Genet. 2023 May 4;19(5):e1010751. doi: 10.1371/journal.pgen.1010751. eCollection 2023 May. PLoS Genet. 2023. PMID: 37141297 Free PMC article.
-
The dissection of tomato flavor: biochemistry, genetics, and omics.Front Plant Sci. 2023 Jun 6;14:1144113. doi: 10.3389/fpls.2023.1144113. eCollection 2023. Front Plant Sci. 2023. PMID: 37346138 Free PMC article. Review.
-
Bioinformatics analysis of the tomato (Solanum lycopersicum) methylesterase gene family.BMC Plant Biol. 2025 May 16;25(1):649. doi: 10.1186/s12870-025-06625-4. BMC Plant Biol. 2025. PMID: 40380152 Free PMC article.
-
Function and Evolution of the Plant MES Family of Methylesterases.Plants (Basel). 2024 Nov 29;13(23):3364. doi: 10.3390/plants13233364. Plants (Basel). 2024. PMID: 39683156 Free PMC article. Review.
References
-
- Agricultural Marketing Resource Center (2018) https://www.agmrc.org/commodities-products/vegetables/tomatoes
-
- Bailey T, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 28–36 - PubMed
-
- Bradbury P, Zhang Z, Kroon D, Casstevens T, Ramdoss Y, Buckler E (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

