RNA polymerase II transcription attenuation at the yeast DNA repair gene DEF1 is biologically significant and dependent on the Hrp1 RNA-recognition motif
- PMID: 36315099
- PMCID: PMC9836349
- DOI: 10.1093/g3journal/jkac292
RNA polymerase II transcription attenuation at the yeast DNA repair gene DEF1 is biologically significant and dependent on the Hrp1 RNA-recognition motif
Abstract
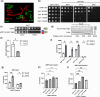
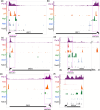
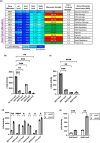
Premature transcription termination (i.e. attenuation) is a potent gene regulatory mechanism that represses mRNA synthesis. Attenuation of RNA polymerase II is more prevalent than once appreciated, targeting 10-15% of mRNA genes in yeast through higher eukaryotes, but its significance and mechanism remain obscure. In the yeast Saccharomyces cerevisiae, polymerase II attenuation was initially shown to rely on Nrd1-Nab3-Sen1 termination, but more recently our laboratory characterized a hybrid termination pathway involving Hrp1, an RNA-binding protein in the 3'-end cleavage factor. One of the hybrid attenuation gene targets is DEF1, which encodes a repair protein that promotes degradation of polymerase II stalled at DNA lesions. In this study, we characterized the chromosomal DEF1 attenuator and the functional role of Hrp1. DEF1 attenuator mutants overexpressed Def1 mRNA and protein, exacerbated polymerase II degradation, and hindered cell growth, supporting a biologically significant DEF1 attenuator function. Using an auxin-induced Hrp1 depletion system, we identified new Hrp1-dependent attenuators in MNR2, SNG1, and RAD3 genes. An hrp1-5 mutant (L205S) known to impair binding to cleavage factor protein Rna14 also disrupted attenuation, but surprisingly no widespread defect was observed for an hrp1-1 mutant (K160E) located in the RNA-recognition motif. We designed a new RNA recognition motif mutant (hrp1-F162W) that altered a highly conserved residue and was lethal in single copy. In a heterozygous strain, hrp1-F162W exhibited dominant-negative readthrough defects at several gene attenuators. Overall, our results expand the hybrid RNA polymerase II termination pathway, confirming that Hrp1-dependent attenuation controls multiple yeast genes and may function through binding cleavage factor proteins and/or RNA.
Keywords: DEF1; CPF-CF; Hrp1; NNS; RNA polymerase II; RRM; attenuation; transcription termination.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Domains and residues of the Saccharomyces cerevisiae hnRNP protein Hrp1 important for transcriptional autoregulation and noncoding RNA termination.Genetics. 2023 Aug 31;225(1):iyad134. doi: 10.1093/genetics/iyad134. Genetics. 2023. PMID: 37467478 Free PMC article.
-
RNA Polymerase II Transcription Attenuation at the Yeast DNA Repair Gene, DEF1, Involves Sen1-Dependent and Polyadenylation Site-Dependent Termination.G3 (Bethesda). 2018 May 31;8(6):2043-2058. doi: 10.1534/g3.118.200072. G3 (Bethesda). 2018. PMID: 29686108 Free PMC article.
-
Transcriptomes of six mutants in the Sen1 pathway reveal combinatorial control of transcription termination across the Saccharomyces cerevisiae genome.PLoS Genet. 2017 Jun 30;13(6):e1006863. doi: 10.1371/journal.pgen.1006863. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28665995 Free PMC article.
-
The Nrd1-Nab3-Sen1 transcription termination complex from a structural perspective.Biochem Soc Trans. 2023 Jun 28;51(3):1257-1269. doi: 10.1042/BST20221418. Biochem Soc Trans. 2023. PMID: 37222282 Free PMC article. Review.
-
Transcription termination and the control of the transcriptome: why, where and how to stop.Nat Rev Mol Cell Biol. 2015 Mar;16(3):190-202. doi: 10.1038/nrm3943. Epub 2015 Feb 4. Nat Rev Mol Cell Biol. 2015. PMID: 25650800 Review.
Cited by
-
Domains and residues of the Saccharomyces cerevisiae hnRNP protein Hrp1 important for transcriptional autoregulation and noncoding RNA termination.Genetics. 2023 Aug 31;225(1):iyad134. doi: 10.1093/genetics/iyad134. Genetics. 2023. PMID: 37467478 Free PMC article.
References
-
- Arigo JT, Carroll KL, Ames JM, Corden JL.. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell. 2006;21(5):641–651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
