Integrated analysis of lncRNA and gene expression in longissimus dorsi muscle at two developmental stages of Hainan black goats
- PMID: 36315512
- PMCID: PMC9621442
- DOI: 10.1371/journal.pone.0276004
Integrated analysis of lncRNA and gene expression in longissimus dorsi muscle at two developmental stages of Hainan black goats
Abstract
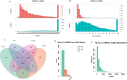
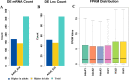
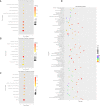
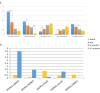
It is deemed that meat quality of kids' is better than that of adults' for Hainan black goat. Generally, meat quality is affected by many indicators, such as intramuscular fat (IMF) content, muscle fiber diameter and shear force. It is indicated that long non-coding RNAs (lncRNAs) play essential roles in meat quality of goats. However, it is unclear whether and how lncRNAs and genes play their roles in meat quality of Hainan Black goats. Here, we firstly compared the meat quality between two-month-old kids (kids) and adult goats (adults). Then, the lncRNA-seq and RNA-seq data were integrated and analyzed to explore the potential functions of lncRNAs and genes. The results showed that adults' IMF content and muscle fiber diameter were extremely significantly higher than that of kids (P<0.01). For the sequenced data, average 84,970,398, and 83,691,250 clean reads were obtained respectively for Kids and adults, among which ~96% were mapped to the reference genome of goats. Through analyzing, 18,242 goat annotated genes, 1,429 goat annotated lncRNAs and 2,967 novel lncRNAs were obtained. Analysis of differential expression genes (DEGs) and lncRNAs (DELs) showed that 328 DEGs and 98 DELs existed between kids and adults. Furthermore, functional enrichment analysis revealed that a number of DEGs and DELs were mainly associated with IMF. Primarily, DGAT2 expressed higher in adults than that in kids and CPT1A expressed higher in kids than that in adults. Both of them were overlapped by DEGs and targets of DELs, suggesting the two DEGs and the DELs targeted by the two DEGs might be the potential regulators of goat IMF deposition. Taken together, our results provide basic support for further understanding the function and mechanism of lncRNAs and genes in meat quality of Hainan black goats.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Identification of differentially expressed long non-coding RNAs and messenger RNAs involved with muscle development in Dazu black goats through RNA sequencing.Anim Biotechnol. 2023 Nov;34(4):1305-1313. doi: 10.1080/10495398.2021.2020804. Epub 2022 Jan 5. Anim Biotechnol. 2023. PMID: 34985384
-
Integrated transcriptome analysis reveals roles of long non-coding RNAs (lncRNAs) in caprine skeletal muscle mass and meat quality.Funct Integr Genomics. 2023 Feb 22;23(1):63. doi: 10.1007/s10142-023-00987-4. Funct Integr Genomics. 2023. PMID: 36810929
-
Co-expression analysis of long non-coding RNAs and mRNAs involved in intramuscular fat deposition in Muchuan black-bone chicken.Br Poult Sci. 2023 Jun;64(3):289-298. doi: 10.1080/00071668.2022.2162370. Epub 2023 Feb 22. Br Poult Sci. 2023. PMID: 36622203
-
Long-stranded non-coding RNAs temporal-specific expression profiles reveal longissimus dorsi muscle development and intramuscular fat deposition in Tianzhu white yak.J Anim Sci. 2023 Jan 3;101:skad394. doi: 10.1093/jas/skad394. J Anim Sci. 2023. PMID: 38029315 Free PMC article.
-
Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production.Cells. 2019 Sep 19;8(9):1107. doi: 10.3390/cells8091107. Cells. 2019. PMID: 31546877 Free PMC article. Review.
Cited by
-
One Copy Number Variation within the Angiopoietin-1 Gene Is Associated with Leizhou Black Goat Meat Quality.Animals (Basel). 2024 Sep 14;14(18):2682. doi: 10.3390/ani14182682. Animals (Basel). 2024. PMID: 39335271 Free PMC article.
-
Long non-coding RNA (LncRNA) and epigenetic factors: their role in regulating the adipocytes in bovine.Front Genet. 2024 Oct 3;15:1405588. doi: 10.3389/fgene.2024.1405588. eCollection 2024. Front Genet. 2024. PMID: 39421300 Free PMC article. Review.
-
Analysis of lncRNAs and Their Regulatory Network in Skeletal Muscle Development of the Yangtze River Delta White Goat.Animals (Basel). 2024 Oct 30;14(21):3125. doi: 10.3390/ani14213125. Animals (Basel). 2024. PMID: 39518848 Free PMC article.
-
Integrated TWAS, GWAS, and RNAseq results identify candidate genes associated with reproductive traits in cows.Sci Rep. 2025 Jan 14;15(1):1932. doi: 10.1038/s41598-024-82448-x. Sci Rep. 2025. PMID: 39809816 Free PMC article.
-
Effects of Copy Number Variations in the Plectin (PLEC) Gene on the Growth Traits and Meat Quality of Leizhou Black Goats.Animals (Basel). 2023 Nov 25;13(23):3651. doi: 10.3390/ani13233651. Animals (Basel). 2023. PMID: 38067002 Free PMC article.
References
-
- Liu Y, Wang SL, Zhang JF, Zhang W, Li W. LncRNA-disease associations prediction based on neural network-based matrix factorization. IEEE Access. 2020;PP(99):1–9.
-
- Ma Y, Liu Y, Pu YS, Cui ML, Mao ZJ, Li ZZ, et al.. LncRNA IGFL2-AS1 functions as a ceRNA in regulating ARPP19 through competitive binding to miR2 in gastric cancer. Mol Carcinog. 2020;59(3):311–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

