Hereditary variants of unknown significance in African American women with breast cancer
- PMID: 36315513
- PMCID: PMC9621418
- DOI: 10.1371/journal.pone.0273835
Hereditary variants of unknown significance in African American women with breast cancer
Abstract
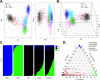
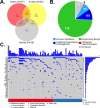
Expanded implementation of genetic sequencing has precipitously increased the discovery of germline and somatic variants. The direct benefit of identifying variants in actionable genes may lead to risk reduction strategies such as increased surveillance, prophylactic surgery, as well as lifestyle modifications to reduce morbidity and mortality. However, patients with African ancestry are more likely to receive inconclusive genetic testing results due to an increased number of variants of unknown significance decreasing the utility and impact on disease management and prevention. This study examines whole exome sequencing results from germline DNA samples in African American women with a family history of cancer including 37 cases that were diagnosed with breast cancer and 51 family members. Self-identified ancestry was validated and compared to the 1000 genomes population. The analysis of sequencing results was limited to 85 genes from three clinically available common genetic screening platforms. This target region had a total of 993 variants of which 6 (<1%) were pathogenic or likely pathogenic, 736 (74.1%) were benign, and 170 (17.1%) were classified as a variant of unknown significance. There was an average of 3.4±1.8 variants with an unknown significance per individual and 85 of 88 individuals (96.6%) harbored at least one of these in the targeted genes. Pathogenic or likely pathogenic variants were only found in 6 individuals for the BRCA1 (p.R1726fs, rs80357867), BRCA2 (p.K589fs, rs397507606 & p.L2805fs, rs397507402), RAD50 (p.E995fs, rs587780154), ATM (p.V2424G, rs28904921), or MUTYH (p.G396D, rs36053993) genes. Strategies to functionally validate the remaining variants of unknown significance, especially in understudied and hereditary cancer populations, are greatly needed to increase the clinical utility and utilization of clinical genetic screening platforms to reduce cancer incidence and mortality.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

