Dynamic alternative DNA structures in biology and disease
- PMID: 36316397
- PMCID: PMC11634456
- DOI: 10.1038/s41576-022-00539-9
Dynamic alternative DNA structures in biology and disease
Abstract
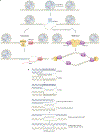
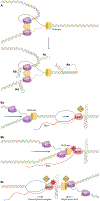
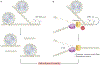
Repetitive elements in the human genome, once considered 'junk DNA', are now known to adopt more than a dozen alternative (that is, non-B) DNA structures, such as self-annealed hairpins, left-handed Z-DNA, three-stranded triplexes (H-DNA) or four-stranded guanine quadruplex structures (G4 DNA). These dynamic conformations can act as functional genomic elements involved in DNA replication and transcription, chromatin organization and genome stability. In addition, recent studies have revealed a role for these alternative structures in triggering error-generating DNA repair processes, thereby actively enabling genome plasticity. As a driving force for genetic variation, non-B DNA structures thus contribute to both disease aetiology and evolution.
© 2022. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
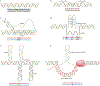

References
-
- Nurk S et al. The complete sequence of a human genome. Science 376, 44–53 (2022). - PMC - PubMed
-
The most recent and compete human genome sequencing assembly reveals more repetitive elements in the human genome than researchers have previously estimated, which could potentially support non-B DNA formation.
-
- Plohl M, Luchetti A, Mestrovic N & Mantovani B Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409, 72–82 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

