Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing
- PMID: 36316451
- PMCID: PMC9712115
- DOI: 10.1038/s41564-022-01258-x
Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing
Abstract
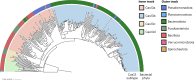
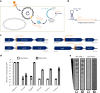
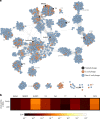
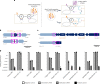
CRISPR-Cas13 proteins are RNA-guided RNA nucleases that defend against incoming RNA and DNA phages by binding to complementary target phage transcripts followed by general, non-specific RNA degradation. Here we analysed the defensive capabilities of LbuCas13a from Leptotrichia buccalis and found it to have robust antiviral activity unaffected by target phage gene essentiality, gene expression timing or target sequence location. Furthermore, we find LbuCas13a antiviral activity to be broadly effective against a wide range of phages by challenging LbuCas13a against nine E. coli phages from diverse phylogenetic groups. Leveraging the versatility and potency enabled by LbuCas13a targeting, we applied LbuCas13a towards broad-spectrum phage editing. Using a two-step phage-editing and enrichment method, we achieved seven markerless genome edits in three diverse phages with 100% efficiency, including edits as large as multi-gene deletions and as small as replacing a single codon. Cas13a can be applied as a generalizable tool for editing the most abundant and diverse biological entities on Earth.
© 2022. The Author(s).
Conflict of interest statement
J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics and Mammoth Biosciences; a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, Algen Biotechnologies, Felix Biosciences, The Column Group and Inari; a director at Johnson & Johnson and Tempus; and has research projects sponsored by Biogen, Pfizer, AppleTree Partners and Roche. J.B. is a founder of Metagenomi. R.B. is a shareholder of Caribou Biosciences, Intellia Therapeutics, Locus Biosciences, Inari, TreeCo and Ancilia Biosciences. V.K.M. is a co-founder of Felix Biotechnology. All other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

