Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses
- PMID: 36316475
- PMCID: PMC9869360
- DOI: 10.1038/s41590-022-01328-6
Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses
Abstract
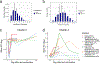
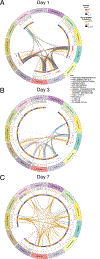
Systems vaccinology has defined molecular signatures and mechanisms of immunity to vaccination. However, comparative analysis of immunity to different vaccines is lacking. We integrated transcriptional data of over 3,000 samples, from 820 adults across 28 studies of 13 vaccines and analyzed vaccination-induced signatures of antibody responses. Most vaccines induced signatures of innate immunity and plasmablasts at days 1 and 7, respectively, after vaccination. However, the yellow fever vaccine induced an early transient signature of T and B cell activation at day 1, followed by delayed antiviral/interferon and plasmablast signatures that peaked at days 7 and 14-21, respectively. Thus, there was no evidence for a 'universal signature' that predicted antibody response to all vaccines. However, accounting for the asynchronous nature of responses, we defined a time-adjusted signature that predicted antibody responses across vaccines. These results provide a transcriptional atlas of immunity to vaccination and define a common, time-adjusted signature of antibody responses.
© 2022. Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
OL is a named inventor on patents held by Boston Children’s Hospital regarding human
Figures









Comment in
-
Immune signature atlas of vaccines: learning from the good responders.Nat Immunol. 2022 Dec;23(12):1654-1656. doi: 10.1038/s41590-022-01361-5. Nat Immunol. 2022. PMID: 36443516 Free PMC article.
References
-
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–25. - PMC - PubMed
-
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR 3rd, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sekaly RP. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–31. - PMC - PubMed
-
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–95. - PMC - PubMed
Methods References:
-
- Irizarry RA et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). - PubMed
-
- Dewey M metap: meta-analysis of significance values. R package version 1.8; 2022.
Publication types
MeSH terms
Substances
Grants and funding
- U19 AI128913/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U19 AI057266/AI/NIAID NIH HHS/United States
- R37 DK057665/DK/NIDDK NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- U01 AI165527/AI/NIAID NIH HHS/United States
- U19 AI128914/AI/NIAID NIH HHS/United States
- U19 AI118626/AI/NIAID NIH HHS/United States
- U19 AI167903/AI/NIAID NIH HHS/United States
- U19 AI128949/AI/NIAID NIH HHS/United States
- U19 AI090023/AI/NIAID NIH HHS/United States
- R01 AI048638/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

