Evaluation of the use of CRISPR loci for discrimination of Salmonella enterica subsp. enterica serovar Enteritidis strains recovered in Canada and comparison with other subtyping methods
- PMID: 36317002
- PMCID: PMC9576496
- DOI: 10.3934/microbiol.2022022
Evaluation of the use of CRISPR loci for discrimination of Salmonella enterica subsp. enterica serovar Enteritidis strains recovered in Canada and comparison with other subtyping methods
Abstract
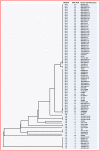
Salmonella enterica subsp. enterica serovar Enteritidis remains one of the most important foodborne pathogens worldwide. To minimise its public health impact when outbreaks of the disease occur, timely investigation to identify and recall the contaminated food source is necessary. Central to this approach is the need for rapid and accurate identification of the bacterial subtype epidemiologically linked to the outbreak. While traditional methods of S. Enteritidis subtyping, such as pulsed field gel electrophoresis (PFGE) and phage typing (PT), have played an important role, the clonal nature of this organism has spurred efforts to improve subtyping resolution and timeliness through molecular based approaches. This study uses a cohort of 92 samples, recovered from a variety of sources, to compare these two traditional methods for S. Enteritidis subtyping with recently developed molecular techniques. These latter methods include the characterisation of two clustered regularly interspaced short palindromic repeats (CRISPR) loci, either in isolation or together with sequence analysis of virulence genes such as fimH. For comparison, another molecular technique developed in this laboratory involved the scoring of 60 informative single nucleotide polymorphisms (SNPs) distributed throughout the genome. Based on both the number of subtypes identified and Simpson's index of diversity, the CRISPR method was the least discriminatory and not significantly improved with the inclusion of fimH gene sequencing. While PT analysis identified the most subtypes, the SNP-PCR process generated the greatest index of diversity value. Combining methods consistently improved the number of subtypes identified, with the SNP/CRISPR typing scheme generating a level of diversity comparable with that of PT/PFGE. While these molecular methods, when combined, may have significant utility in real-world situations, this study suggests that CRISPR analysis alone lacks the discriminatory capability required to support investigations of foodborne disease outbreaks.
Keywords: CRISPR; PFGE; S. Enteritidis; SNP-PCR; Salmonella enterica; phage; serovar; subtyping.
© 2022 the Author(s), licensee AIMS Press.
Conflict of interest statement
Conflict of interest: All authors declare no conflicts of interest in this paper.
Figures


References
-
- PHAC. National Enteric Surveillance Program Annual Summary 2016. Guelph, Ontario: Public Health Agency, Government of Canada; 2018.
LinkOut - more resources
Full Text Sources
