CRISPR-Cas9-induced gene knockout in zebrafish
- PMID: 36317180
- PMCID: PMC9617198
- DOI: 10.1016/j.xpro.2022.101779
CRISPR-Cas9-induced gene knockout in zebrafish
Abstract
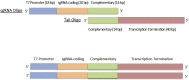
The application of CRISPR has greatly facilitated genotype-phenotype studies of human disease models. In this protocol, we describe CRISPR-Cas9-induced gene knockout in zebrafish, utilizing purified Cas9 protein and in vitro-transcribed sgRNA. This protocol targets the PHLPP1 gene in an Indian wild-caught strain, but is broadly applicable. Major factors influencing protocol success include zebrafish health and fecundity, sgRNA efficiency and specificity, germline transmission, and mutant viability. For complete details on the use and execution of this protocol, please refer to Balamurugan et al. (2022).
Keywords: CRISPR; Genetics; Model organisms; Molecular biology; Sequencing.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Balamurugan K., Medishetti R., Kotha J., Behera P., Chandra K., Mavuduru V.A., Joshi M.B., Samineni R., Katika M.R., Ball W.B., et al. PHLPP1 promotes neutral lipid accumulation through AMPK/ChREBP-dependent lipid uptake and fatty acid synthesis pathways. iScience. 2022;25:103766. doi: 10.1016/j.isci.2022.103766. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

