Unraveling Complex Local Protein Environments with 4-Cyano-l-phenylalanine
- PMID: 36317866
- PMCID: PMC10234312
- DOI: 10.1021/acs.jpcb.2c05954
Unraveling Complex Local Protein Environments with 4-Cyano-l-phenylalanine
Abstract
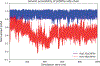
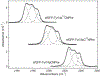
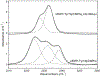
We present a multifaceted approach to effectively probe complex local protein environments utilizing the vibrational reporter unnatural amino acid (UAA) 4-cyano-l-phenylalanine (pCNPhe) in the model system superfolder green fluorescent protein (sfGFP). This approach combines temperature-dependent infrared (IR) spectroscopy, X-ray crystallography, and molecular dynamics (MD) simulations to provide a molecular interpretation of the local environment of the nitrile group in the protein. Specifically, a two-step enantioselective synthesis was developed that provided an 87% overall yield of pCNPhe in high purity without the need for chromatography. It was then genetically incorporated individually at three unique sites (74, 133, and 149) in sfGFP to probe these local protein environments. The incorporation of the UAA site-specifically in sfGFP utilized an engineered, orthogonal tRNA synthetase in E. coli using the Amber codon suppression protocol, and the resulting UAA-containing sfGFP constructs were then explored with this approach. This methodology was effectively utilized to further probe the local environments of two surface sites (sites 133 and 149) that we previously explored with room temperature IR spectroscopy and X-ray crystallography and a new interior site (site 74) featuring a complex local environment around the nitrile group of pCNPhe. Site 133 was found to be solvent-exposed, while site 149 was partially buried. Site 74 was found to consist of three distinct local environments around the nitrile group including nonspecific van der Waals interactions, hydrogen-bonding to a structural water, and hydrogen-bonding to a histidine side chain.
Figures


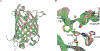
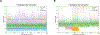
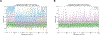
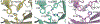
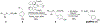
References
-
- Getahun Z; Huang C-Y; Wang T; De León B; DeGrado WF; Gai F J. Am. Chem. Soc. 2003, 125, 405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

