The Neuroscience Multi-Omic Archive: a BRAIN Initiative resource for single-cell transcriptomic and epigenomic data from the mammalian brain
- PMID: 36318260
- PMCID: PMC9825473
- DOI: 10.1093/nar/gkac962
The Neuroscience Multi-Omic Archive: a BRAIN Initiative resource for single-cell transcriptomic and epigenomic data from the mammalian brain
Abstract
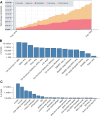
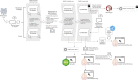
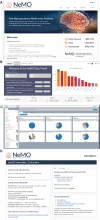
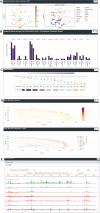
Scalable technologies to sequence the transcriptomes and epigenomes of single cells are transforming our understanding of cell types and cell states. The Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative Cell Census Network (BICCN) is applying these technologies at unprecedented scale to map the cell types in the mammalian brain. In an effort to increase data FAIRness (Findable, Accessible, Interoperable, Reusable), the NIH has established repositories to make data generated by the BICCN and related BRAIN Initiative projects accessible to the broader research community. Here, we describe the Neuroscience Multi-Omic Archive (NeMO Archive; nemoarchive.org), which serves as the primary repository for genomics data from the BRAIN Initiative. Working closely with other BRAIN Initiative researchers, we have organized these data into a continually expanding, curated repository, which contains transcriptomic and epigenomic data from over 50 million brain cells, including single-cell genomic data from all of the major regions of the adult and prenatal human and mouse brains, as well as substantial single-cell genomic data from non-human primates. We make available several tools for accessing these data, including a searchable web portal, a cloud-computing interface for large-scale data processing (implemented on Terra, terra.bio), and a visualization and analysis platform, NeMO Analytics (nemoanalytics.org).
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Zeng H., Sanes J.R.. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 2017; 18:530–546. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

