KLF9 and KLF13 transcription factors boost myelin gene expression in oligodendrocytes as partners of SOX10 and MYRF
- PMID: 36318265
- PMCID: PMC9723594
- DOI: 10.1093/nar/gkac953
KLF9 and KLF13 transcription factors boost myelin gene expression in oligodendrocytes as partners of SOX10 and MYRF
Abstract
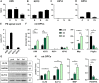
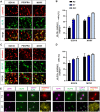
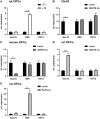
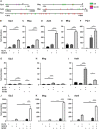
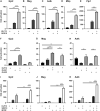
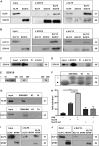
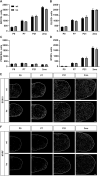
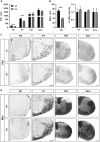
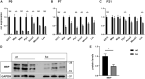
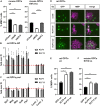
Differentiated oligodendrocytes produce myelin and thereby ensure rapid nerve impulse conduction and efficient information processing in the vertebrate central nervous system. The Krüppel-like transcription factor KLF9 enhances oligodendrocyte differentiation in culture, but appears dispensable in vivo. Its mode of action and role within the oligodendroglial gene regulatory network are unclear. Here we show that KLF9 shares its expression in differentiating oligodendrocytes with the closely related KLF13 protein. Both KLF9 and KLF13 bind to regulatory regions of genes that are important for oligodendrocyte differentiation and equally recognized by the central differentiation promoting transcription factors SOX10 and MYRF. KLF9 and KLF13 physically interact and synergistically activate oligodendrocyte-specific regulatory regions with SOX10 and MYRF. Similar to KLF9, KLF13 promotes differentiation and myelination in primary oligodendroglial cultures. Oligodendrocyte differentiation is also altered in KLF13-deficient mice as demonstrated by a transiently reduced myelin gene expression during the first postnatal week. Considering mouse phenotypes, the similarities in expression pattern and genomic binding and the behaviour in functional assays, KLF9 and KLF13 are important and largely redundant components of the gene regulatory network in charge of oligodendrocyte differentiation and myelination.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010; 330:779–782. - PubMed
-
- Sock E., Wegner M.. Transcriptional control of myelination and remyelination. Glia. 2019; 67:2153–2165. - PubMed
-
- Sock E., Wegner M.. Using the lineage determinants olig2 and sox10 to explore transcriptional regulation of oligodendrocyte development. Dev. Neurobiol. 2021; 81:892–901. - PubMed
-
- Nemer M., Horb M.E.. The KLF family of transcriptional regulators in cardiomyocyte proliferation and differentiation. Cell Cycle. 2007; 6:117–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

