A novel application of neural networks to identify potentially effective combinations of biologic factors for enhancement of bone fusion/repair
- PMID: 36318539
- PMCID: PMC9624421
- DOI: 10.1371/journal.pone.0276562
A novel application of neural networks to identify potentially effective combinations of biologic factors for enhancement of bone fusion/repair
Abstract
Introduction: The use of biologic adjuvants (orthobiologics) is becoming commonplace in orthopaedic surgery. Among other applications, biologics are often added to enhance fusion rates in spinal surgery and to promote bone healing in complex fracture patterns. Generally, orthopaedic surgeons use only one biomolecular agent (ie allograft with embedded bone morphogenic protein-2) rather than several agents acting in concert. Bone fusion, however, is a highly multifactorial process and it likely could be more effectively enhanced using biologic factors in combination, acting synergistically. We used artificial neural networks, trained via machine learning on experimental data on orthobiologic interventions and their outcomes, to identify combinations of orthobiologic factors that potentially would be more effective than single agents. This use of machine learning applied to orthobiologic interventions is unprecedented.
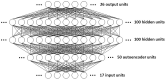
Methods: Available data on the outcomes associated with various orthopaedic biologic agents, electrical stimulation, and pulsed ultrasound were curated from the literature and assembled into a form suitable for machine learning. The best among many different types of neural networks was chosen for its ability to generalize over this dataset, and that network was used to make predictions concerning the expected efficacy of 2400 medically feasible combinations of 9 different agents and treatments.
Results: The most effective combinations were high in the bone-morphogenic proteins (BMP) 2 and 7 (BMP2, 15mg; BMP7, 5mg), and in osteogenin (150ug). In some of the most effective combinations, electrical stimulation could substitute for osteogenin. Some other effective combinations also included bone marrow aspirate concentrate. BMP2 and BMP7 appear to have the strongest pairwise linkage of the factors analyzed in this study.
Conclusions: Artificial neural networks are powerful forms of artificial intelligence that can be applied readily in the orthopaedic domain, but neural network predictions improve along with the amount of data available to train them. This study provides a starting point from which networks trained on future, expanded datasets can be developed. Yet even this initial model makes specific predictions concerning potentially effective combinatorial therapeutics that should be verified experimentally. Furthermore, our analysis provides an avenue for further research into the basic science of bone healing by demonstrating agents that appear to be linked in function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Goodfellow I, Bengio Y., and Courville A. Deep Learning: MIS Press; 2016.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

