Deciphering molecular and cellular ex vivo responses to bispecific antibodies PD1-TIM3 and PD1-LAG3 in human tumors
- PMID: 36319064
- PMCID: PMC9628669
- DOI: 10.1136/jitc-2022-005548
Deciphering molecular and cellular ex vivo responses to bispecific antibodies PD1-TIM3 and PD1-LAG3 in human tumors
Abstract
Background: Next-generation cancer immunotherapies are designed to broaden the therapeutic repertoire by targeting new immune checkpoints including lymphocyte-activation gene 3 (LAG-3) and T cell immunoglobulin and mucin-domain containing-3 (TIM-3). Yet, the molecular and cellular mechanisms by which either receptor functions to mediate its inhibitory effects are still poorly understood. Similarly, little is known on the differential effects of dual, compared with single, checkpoint inhibition.
Methods: We here performed in-depth characterization, including multicolor flow cytometry, single cell RNA sequencing and multiplex supernatant analysis, using tumor single cell suspensions from patients with cancer treated ex vivo with novel bispecific antibodies targeting programmed cell death protein 1 (PD-1) and TIM-3 (PD1-TIM3), PD-1 and LAG-3 (PD1-LAG3), or with anti-PD-1.
Results: We identified patient samples which were responsive to PD1-TIM3, PD1-LAG3 or anti-PD-1 using an in vitro approach, validated by the analysis of 659 soluble proteins and enrichment for an anti-PD-1 responder signature. We found increased abundance of an activated (HLA-DR+CD25+GranzymeB+) CD8+ T cell subset and of proliferating CD8+ T cells, in response to bispecific antibody or anti-PD-1 treatment. Bispecific antibodies, but not anti-PD-1, significantly increased the abundance of a proliferating natural killer cell subset, which exhibited enrichment for a tissue-residency signature. Key phenotypic and transcriptional changes occurred in a PD-1+CXCL13+CD4+ T cell subset, in response to all treatments, including increased interleukin-17 secretion and signaling toward plasma cells. Interestingly, LAG-3 protein upregulation was detected as a unique pharmacodynamic effect mediated by PD1-LAG3, but not by PD1-TIM3 or anti-PD-1.
Conclusions: Our in vitro system reliably assessed responses to bispecific antibodies co-targeting PD-1 together with LAG-3 or TIM-3 using patients' tumor infiltrating immune cells and revealed transcriptional and phenotypic imprinting by bispecific antibody formats currently tested in early clinical trials.
Keywords: biomarkers, tumor; clinical trials as topic; immunotherapy.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: KH, PG, IID, FJ, DM, AZw, PW, SS, PU, CK, LC-D and HK are current or former employees of F. Hoffmann-La Roche and declare patent applications and stock ownership with Roche. AZ received consulting/advisor fees from Bristol-Myers Squibb, Merck Sharp & Dohme, Hoffmann-La Roche, NBE Therapeutics, Secarna, ACM Pharma and Hookipa and maintains further non-commercial research agreements with Secarna, Hookipa, Roche and Beyondsprings. All other authors declare they have no competing interests.
Figures
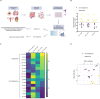




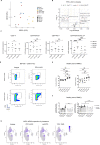
References
-
- Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. 10.1158/2159-8290.CD-18-0367 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
