Identification of quantitative trait loci for related traits of stalk lodging resistance using genome-wide association studies in maize (Zea mays L.)
- PMID: 36319954
- PMCID: PMC9623923
- DOI: 10.1186/s12863-022-01091-5
Identification of quantitative trait loci for related traits of stalk lodging resistance using genome-wide association studies in maize (Zea mays L.)
Abstract
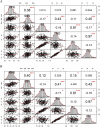
Background: Stalk lodging is one of the main factors affecting maize (Zea mays L.) yield and limiting mechanized harvesting. Developing maize varieties with high stalk lodging resistance requires exploring the genetic basis of lodging resistance-associated agronomic traits. Stalk strength is an important indicator to evaluate maize lodging and can be evaluated by measuring stalk rind penetrometer resistance (RPR) and stalk buckling strength (SBS). Along with morphological traits of the stalk for the third internodes length (TIL), fourth internode length (FIL), third internode diameter (TID), and the fourth internode diameter (FID) traits are associated with stalk lodging resistance.
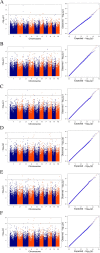
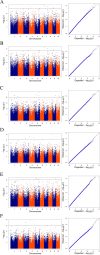
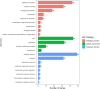
Results: In this study, a natural population containing 248 diverse maize inbred lines genotyped with 83,057 single nucleotide polymorphism (SNP) markers was used for genome-wide association study (GWAS) for six stalk lodging resistance-related traits. The heritability of all traits ranged from 0.59 to 0.72 in the association mapping panel. A total of 85 significant SNPs were identified for the association mapping panel using best linear unbiased prediction (BLUP) values of all traits. Additionally, five candidate genes were associated with stalk strength traits, which were either directly or indirectly associated with cell wall components.
Conclusions: These findings contribute to our understanding of the genetic basis of maize stalk lodging and provide valuable theoretical guidance for lodging resistance in maize breeding in the future.
Keywords: Candidate gene; Genome-wide association study; Maize; Quantitative trait nucleotides; Stalk lodging resistance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
The authors declare that they have no conflicts of interest.
Figures


Similar articles
-
Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize.Int J Mol Sci. 2024 Feb 5;25(3):1918. doi: 10.3390/ijms25031918. Int J Mol Sci. 2024. PMID: 38339196 Free PMC article. Review.
-
Genome-wide association study for stalk lodging resistance related traits in maize (Zea mays L.).BMC Genomics. 2024 Jan 2;25(1):19. doi: 10.1186/s12864-023-09917-x. BMC Genomics. 2024. PMID: 38166629 Free PMC article.
-
Genetic dissection of stalk lodging-related traits using an IBM Syn10 DH population in maize across three environments (Zea mays L.).Mol Genet Genomics. 2019 Oct;294(5):1277-1288. doi: 10.1007/s00438-019-01576-6. Epub 2019 May 28. Mol Genet Genomics. 2019. PMID: 31139941
-
Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population.Genes (Basel). 2022 Mar 24;13(4):579. doi: 10.3390/genes13040579. Genes (Basel). 2022. PMID: 35456384 Free PMC article.
-
Maize and heat stress: Physiological, genetic, and molecular insights.Plant Genome. 2024 Mar;17(1):e20378. doi: 10.1002/tpg2.20378. Epub 2023 Aug 16. Plant Genome. 2024. PMID: 37587553 Review.
Cited by
-
Differences in Physicochemical Properties of Stems in Oat (Avena sativa L.) Varieties with Distinct Lodging Resistance and Their Regulation of Lodging at Different Planting Densities.Plants (Basel). 2024 Sep 30;13(19):2739. doi: 10.3390/plants13192739. Plants (Basel). 2024. PMID: 39409609 Free PMC article.
-
Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize.Int J Mol Sci. 2024 Feb 5;25(3):1918. doi: 10.3390/ijms25031918. Int J Mol Sci. 2024. PMID: 38339196 Free PMC article. Review.
-
Phenotypic and genome-wide association analyses for nitrogen use efficiency related traits in maize (Zea mays L.) exotic introgression lines.Front Plant Sci. 2023 Oct 9;14:1270166. doi: 10.3389/fpls.2023.1270166. eCollection 2023. Front Plant Sci. 2023. PMID: 37877090 Free PMC article.
-
Genome-wide association study for stalk lodging resistance related traits in maize (Zea mays L.).BMC Genomics. 2024 Jan 2;25(1):19. doi: 10.1186/s12864-023-09917-x. BMC Genomics. 2024. PMID: 38166629 Free PMC article.
-
Variation and interrelationships in the growth, yield, and lodging of oat under different planting densities.PeerJ. 2024 Apr 29;12:e17310. doi: 10.7717/peerj.17310. eCollection 2024. PeerJ. 2024. PMID: 38699188 Free PMC article.
References
-
- Li W, Liu W, Liu L, You M, Liu G, Li B. QTL mapping for wheat flour color with additive, epistatic, and QTL × environmental interaction effects. Sci Agric Sin. 2011;10(5):651–660.
-
- Flint-García SA, Mcmullen MD, Darrah LL. Genetic relationship of stalk strength and ear height in maize. Crop Sci. 2003;43:23–31. doi: 10.2135/cropsci2003.0023. - DOI
-
- Flint-García SA, Jampatong C, Darrah LL, McMullen MD. Quantitative trait locus analysis of stalk strength in four maize populations. Crop Sci. 2003;43:13–22. doi: 10.2135/cropsci2003.0013. - DOI
-
- Duvick DN, Cassman KG. Post–green revolution trends in yield potential of temperate maize in the north-central united states. Crop Sci. 1999;39(6):1622–1630. doi: 10.2135/cropsci1999.3961622x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
