Combining multiple acquisition modes and computational data annotation for structural characterization in traditional Chinese medicine: Miao Nationality medicine Qijiao Shengbai Capsule as a case study
- PMID: 36320242
- PMCID: PMC9520537
- DOI: 10.1039/d2ra04720a
Combining multiple acquisition modes and computational data annotation for structural characterization in traditional Chinese medicine: Miao Nationality medicine Qijiao Shengbai Capsule as a case study
Abstract
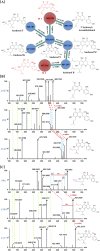
Qijiao Shengbai Capsule (QSC) is a reputable Miao Nationality medicine used for treating leukopenia, but its chemical composition has not yet been elucidated. We herein present a strategy, by integrating multiple data acquisition, computational data annotation and processing methods to visualize and identify the complicated constituents in QSC based on ultra-high-performance liquid chromatography coupled with traveling wave ion mobility quadrupole time-of-flight mass spectrometry (UPLC-TWIMS-QTOF-MS). The multiple data acquisition modes, including data-independent mass spectrometryEnergy (MSE), data-independent high-definition mass spectrometryEnergy (HDMSE), and fast data-dependent acquisition (fast-DDA), in both positive and negative ion modes, were conducted on a Waters-SYNAPT G2-Si mass spectrometer with an ESI source. An in-house library built by the UNIFI platform could efficiently process the peak annotation of known compounds, whilst different structural types were clustered in the molecular networks for the analogous classification and structural annotation of the unknown ones. Neutral loss, diagnostic ions, feature fragmentation behaviors, and community curation of mass spectrometry data of known compounds helped exploit those similar neighboring nodes of unknown compounds. Moreover, by combination of the predicted CCS values from CCS platform with the experimental CCS values from HDMSE, as well as diagnostic fragment ions, isomer compounds were annotated. By integrating reference compound comparison, a total of 202 constituents, including 94 flavonoids, 12 saponins, 30 phthalides, 38 organic acids, 3 amino acids, 7 alkaloids and 18 others, were unambiguously characterized or tentatively identified in QSC. Among them, 5 potential new compounds were detected and 12 pairs of isomers were comprehensively distinguished. Conclusively, the established multiple acquisition modes, computational data processing and analysis strategy proved to be useful for the in-depth structural identification of QSC.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures







References
-
- Tang M. K. Luo L. L. Zhang C. Wu J. H. Wang X. Y. J. Tradit. Chin. Med. Sci. 2021;8:S22–S26.
-
- Li Z. Y. Tu Y. Li H. T. He J. Que S. Dong G. P. Zhang M. S. Liu J. Q. Huang X. L. Wang X. R. Bolat M. Feng X. Zhang F. B. Jiang F. Zhongguo Zhongyao Zazhi. 2020;45:2265–2274. - PubMed
-
- Wu Y. L. Zhao L. Gu L. F. Tilyek A. Yu B. Y. Chai C. Z. J. Ethnopharmacol. 2022;283:114696. - PubMed
-
- Wuniqiemu T. Qin J. J. Teng F. Z. Nabijan M. Cui J. Yi L. Tang W. F. Zhu X. Y. Abduwaki M. Nurahmat M. Wei Y. Dong J. C. J. Ethnopharmacol. 2021;266:113343. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

