Enzyme-linked aptamer-based sandwich assay (ELASA) for detecting Plasmodium falciparum lactate dehydrogenase, a malarial biomarker
- PMID: 36320752
- PMCID: PMC9562052
- DOI: 10.1039/d2ra03796c
Enzyme-linked aptamer-based sandwich assay (ELASA) for detecting Plasmodium falciparum lactate dehydrogenase, a malarial biomarker
Abstract
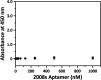
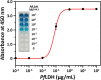
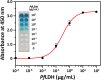
Herein, we report a sensitive and selective enzyme-linked aptamer-based sandwich assay (ELASA) to detect Plasmodium falciparum lactate dehydrogenase (PfLDH), which is an attractive biomarker for malaria diagnosis and antimalarial medication. We performed the sandwich assay with a single aptamer sequence, called 2008s, owing to the structural properties of the PfLDH tetramer instead of using a conventional sandwich assay with two different aptamers (or antibodies) for capturing and probing a target molecule. First, the biotinylated PfLDH aptamer was linked with immobilized streptavidin on a microwell plate for binding flexibility, and then PfLDH was bound to the aptamer. Next, a horseradish peroxidase-conjugated aptamer of the same sequence was used to analyze PfLDH quantitatively. Using this approach, the limit of detection (LOD) of PfLDH with the naked eye was 100 ng mL-1, and the LOD and limit of quantification from the absorbance measurements were 34.9 ng mL-1 and 95.5 ng mL-1, respectively, based on PfLDH spiked blood samples. Our proposed method selectively binds PfLDH, not human lactate dehydrogenase. Therefore, this method may be a valuable tool for diagnosing, monitoring, and quarantining malaria cases easily and rapidly.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
-
- World Health Organization (WHO), World Malaria Report 2021, Geneva, 2021
LinkOut - more resources
Full Text Sources
Other Literature Sources

