One-step asparaginyl endopeptidase (Oa AEP1)-based protein immobilization for single-molecule force spectroscopy
- PMID: 36320890
- PMCID: PMC9533667
- DOI: 10.1039/d2cb00135g
One-step asparaginyl endopeptidase (Oa AEP1)-based protein immobilization for single-molecule force spectroscopy
Abstract
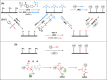
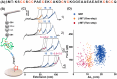
Enzymatic protein ligation has become the most powerful and widely used method for high-precision atomic force microscopy single-molecule force spectroscopy (AFM-SMFS) study of protein mechanics. However, this methodology typically requires the functionalization of the glass surface with a corresponding peptide sequence/tag for enzymatic recognition and multiple steps are needed. Thus, it is time-consuming and a high level of experience is needed for reliable results. To solve this problem, we simplified the procedure using two strategies both based on asparaginyl endopeptidase (AEP). First, we designed a heterobifunctional peptide-based crosslinker, GL-peptide-propargylglycine, which links to an N 3-functionalized surface via the click reaction. Then, the target protein with a C-terminal NGL sequence can be immobilized via the AEP-mediated ligation. Furthermore, we took advantage of the direct ligation between primary amino in a small molecule and protein with C-terminal NGL by AEP. Thus, the target protein can be immobilized on an amino-functionalized surface via AEP in one step. Both approaches were successfully applied to the AFM-SMFS study of eGFP, showing consistent single-molecule results.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Garcia-Galan C. Berenguer-Murcia Á. Fernandez-Lafuente R. Rodrigues R. C. Adv. Synth. Catal. 2011;353:2885–2904. doi: 10.1002/adsc.201100534. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

