Intestinal Apc-inactivation induces HSP25 dependency
- PMID: 36321561
- PMCID: PMC9727927
- DOI: 10.15252/emmm.202216194
Intestinal Apc-inactivation induces HSP25 dependency
Abstract
The majority of colorectal cancers (CRCs) present with early mutations in tumor suppressor gene APC. APC mutations result in oncogenic activation of the Wnt pathway, which is associated with hyperproliferation, cytoskeletal remodeling, and a global increase in mRNA translation. To compensate for the increased biosynthetic demand, cancer cells critically depend on protein chaperones to maintain proteostasis, although their function in CRC remains largely unexplored. In order to investigate the role of molecular chaperones in driving CRC initiation, we captured the transcriptomic profiles of murine wild type and Apc-mutant organoids during active transformation. We discovered a strong transcriptional upregulation of Hspb1, which encodes small heat shock protein 25 (HSP25). We reveal an indispensable role for HSP25 in facilitating Apc-driven transformation, using both in vitro organoid cultures and mouse models, and demonstrate that chemical inhibition of HSP25 using brivudine reduces the development of premalignant adenomas. These findings uncover a hitherto unknown vulnerability in intestinal transformation that could be exploited for the development of chemopreventive strategies in high-risk individuals.
Keywords: Apc mutations; Wnt signaling; colorectal cancer; heat shock proteins; intestinal stem cells.
© 2022 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

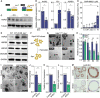
- A
Schematic illustration of the in vitro recombination experiment. TAM, 4OH‐tamoxifen.
- B
Representative phase images of the morphology of WT and Apc −/− organoids before mRNA isolation. Scalebar, 250 μm.
- C
Heat map illustrating differentially expressed genes (DEGs) encoding heat shock proteins between WT and Apc −/− organoids.
- D
Volcano plot of DEGs between normal mouse intestinal tissue and adenomas (GSE65461).
- E, F
Relative gene expression of Hspb1 (***P = 0.0004, n = 6) (E) and protein expression of HSP25 (F) in unrecombined versus recombined murine organoids.
- G
Gene expression of Hspb1 in normal (N) versus adenoma (A) tissue (***P = 0.0002, n = 4).
- H
RNA‐ISH for Hspb1 in murine intestinal adenoma. Scalebar, 100 μm. Zoom panel, 25 μm.
- I
Relative gene expression of human HSPB1 in paired sets of normal versus adenoma tissue, (****P < 0.0001, paired t‐test, n = 59 pairs).
- A
Schematic illustration of the doxycycline‐inducible dnTCF construct.
- B, C
Relative gene expression of AXIN2 (*P = 0.0129), P21 (*P = 0.0103), and HSPB1(*P = 0.0236) (B), and protein expression of HSP27 (C) in Ls174t‐dnTCF4 cells in the presence or absence of doxycycline.
- D, E
Relative gene expression of Hspb1 in murine WT organoids treated with different concentrations of CHIR‐99021 (**P = 0.0016, one way ANOVA, n = 4) (D), and protein expression of HSP25 in WT and Apc −/− organoids (E).
- F
Schematic illustration of in vitro recombination experiment using Hspb1 KO clones.
- G, H
Representative images of single cell Hspb1 KO clones passaged after loss of Apc (G), and quantification of their clonogenic potential (*P = 0.0147(#1), **P = 0.0035(#2), **P = 0.0032(#3)) (H). Scale bar, 500 μm.
- I, J
Representative images of Apc −/− organoids cultured in the absence or presence of 60 μM BVDU (I), and quantification of their clonogenic potential (**P = 0.0039) (J). Scale bar, 500 μm.
- K, L
Relative gene expression of Wnt target genes Axin2 (**P = 0.0080) and Lgr5 (*P = 0.0142) in Apc −/− organoids cultured in the absence or presence of 60 μM BVDU.
- M
RNA‐ISH for Lgr5 in control or BVDU‐treated Apc −/− organoids. Scale bar, 50 μm, zoom panel 20 μm.

- A–C
Gene Set Enrichment Analysis for pathways involved in HSF1‐mediated heat shock response (A), chaperonin‐mediated protein folding (B), and p38/MAPK stress response (C). NES, Normalized Enrichment Score; P, nominal P‐value of the enrichment score, which is based on a phenotype‐based permutation test procedure as described in more detail in (Subramanian et al, 2005).
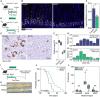
- A
Experimental set‐up of short‐term assay using Villin‐Cre ERT2 ;Apc fl/fl mice. p.o., per os, orally.
- B, C
Visualization of proliferating cells in tissues incubated with EdU for 2‐h (B), and quantification of the hyperproliferative zone (**P = 0.0057, n = 3 mice) (C). Scalebar, 50 μm.
- D
Experimental set‐up of short‐term assay using Lgr5‐Cre ERT2 ;Apc fl/fl mice.
- E, F
RNA‐ISH for Notum reveals Apc‐mutant clones in crypt bottoms of control or BVDU‐treated mice (E), and quantification of the abundance of mutant crypts (n = 2 mice per condition, n = 5 technical replicates per mouse) (F). Scalebar, 50 μm.
- G, H
Clone size distributions of Notum + crypts in control (n = 265 crypts) or BVDU‐treated (n = 260 crypts) mice, the average clone size and number of fixed clones are included in the figure.
- I
Experimental set‐up of tumor development assay using Lgr5‐Cre ERT2 ;Apc fl/fl mice, mice were sacrificed once they develop signs of adenoma development.
- J–L
Survival curves for control and BVDU‐treated mice (P = 0.0351, Mantel‐Cox test) (J), macroscopic images of adenoma development in the distal small intestine (K), and boxplots for the number of adenomas per intestinal region (**P = 0.0013) (L), n = 8 control, n = 11 treated.
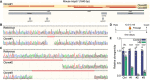
- A
Overview of Hspb1 gene, positions of the sgRNA's and edited sites within the KO clones.
- B, C
Sanger sequencing results of the edited regions in exon 1 (B) or exon 2 (C).
- D, E
Illustration of the heat shock (HS) clonogenicity experiment (D), and quantification of clonogenicity in KO clones in untreated or HS‐treated conditions (****P < 0.0001 for KO#1, 2, and 3, n = 4 experiments, unpaired two‐sided t‐test).

- A
Growth curves of Apc‐mutant organoids.
- B–D
Growth curves of WT organoids (B), representative images of WT organoids cultured in the absence or presence of 60 μM BVDU (C), and quantification of their clonogenic potential (D). Scale bar, 500 μm.
- E
RNA‐ISH for Lgr5 in control or BVDU‐treated WT organoids. Scale bar, 50 μm, zoom panel 20 μm.
- F, G
Representative images (F) and clonogenicity (G) of WT organoids treated with CHIR99021 in the absence or presence of BVDU. (****P < 0.0001, n = 4). Scale bar, 500 μm.
- H, I
Relative Hspb1 (H) and HSP25 (I) expression in wild‐type organoids after heat shock (HS) treatment (**P = 0.0016).
- J–L
Illustration of the heat shock (HS) clonogenicity experiment (J), representative images of control or BVDU‐treated WT organoids after HS treatment (K), and quantification of clonogenicity (*P = 0.0128) (L).

- A
Relative HSPB1 expression in normal colon organoids (n = 3) and organoids derived from patients with familial adenomatous polyposis (**P = 0.0013, n = 4, mean ± s.e.m.).
- B, C
representative images of control or BVDU‐treated human organoids (B) and quantification of clonogenicity (C) (***P = 0.0009 (FAP1), ****P < 0.0001 (FAP2), *P = 0.0197 (FAP3), n = 4 wells per line, data are mean ± s.d.) scale bar, 500 μm.
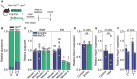
- A
Illustration of experimental setup for crypt isolation.
- B
Ratio of unrecombined versus recombined Apc alleles in control and BVDU treated crypts (n = 2 mice per group, C, control, B, BVDU treated).
- C
Outgrowth of crypts isolated from control and BVDU‐treated mice in ENR (mEGF, Noggin, and Rspondin1) or EN medium (mEGF, Noggin) (**P = 0.0010 (mouse 3), ***P < 0.0001 (mouse 4), n = 4 wells per condition, data are mean ± s.d.).
- D
Relative Notum expression in control or BVDU‐treated Apc organoids (n = 3 experiments).
- E, F
Relative Hspb1 expression in control or tamoxifen‐treated organoids (n = 3 experiments) (E) or intestinal tissues (F) (n = 2 mice per condition, n = 3 technical replicates, C, control, T, Tamoxifen).
References
-
- Bakthisaran R, Tangirala R, Rao CM (2015) Small heat shock proteins: role in cellular functions and pathology. Biochim Biophys Acta 1854: 291–319 - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 - PubMed
-
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H (2009) Crypt stem cells as the cells‐of‐origin of intestinal cancer. Nature 457: 608–611 - PubMed
-
- Benoit Y, Laureys G, Delbeke MJ, De Clercq E (1985) Oral BVDU treatment of varicella and zoster in children with cancer. Eur J Pediatr 143: 198–202 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

