RASCL: Rapid Assessment of Selection in CLades through molecular sequence analysis
- PMID: 36322581
- PMCID: PMC9629619
- DOI: 10.1371/journal.pone.0275623
RASCL: Rapid Assessment of Selection in CLades through molecular sequence analysis
Abstract
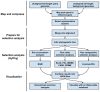
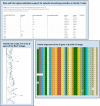
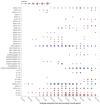
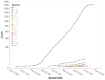
An important unmet need revealed by the COVID-19 pandemic is the near-real-time identification of potentially fitness-altering mutations within rapidly growing SARS-CoV-2 lineages. Although powerful molecular sequence analysis methods are available to detect and characterize patterns of natural selection within modestly sized gene-sequence datasets, the computational complexity of these methods and their sensitivity to sequencing errors render them effectively inapplicable in large-scale genomic surveillance contexts. Motivated by the need to analyze new lineage evolution in near-real time using large numbers of genomes, we developed the Rapid Assessment of Selection within CLades (RASCL) pipeline. RASCL applies state of the art phylogenetic comparative methods to evaluate selective processes acting at individual codon sites and across whole genes. RASCL is scalable and produces automatically updated regular lineage-specific selection analysis reports: even for lineages that include tens or hundreds of thousands of sampled genome sequences. Key to this performance is (i) generation of automatically subsampled high quality datasets of gene/ORF sequences drawn from a selected "query" viral lineage; (ii) contextualization of these query sequences in codon alignments that include high-quality "background" sequences representative of global SARS-CoV-2 diversity; and (iii) the extensive parallelization of a suite of computationally intensive selection analysis tests. Within hours of being deployed to analyze a novel rapidly growing lineage of interest, RASCL will begin yielding JavaScript Object Notation (JSON)-formatted reports that can be either imported into third-party analysis software or explored in standard web-browsers using the premade RASCL interactive data visualization dashboard. By enabling the rapid detection of genome sites evolving under different selective regimes, RASCL is well-suited for near-real-time monitoring of the population-level selective processes that will likely underlie the emergence of future variants of concern in measurably evolving pathogens with extensive genomic surveillance.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures
Update of
-
RASCL: Rapid Assessment Of SARS-CoV-2 Clades Through Molecular Sequence Analysis.bioRxiv [Preprint]. 2022 Jan 18:2022.01.15.476448. doi: 10.1101/2022.01.15.476448. bioRxiv. 2022. Update in: PLoS One. 2022 Nov 2;17(11):e0275623. doi: 10.1371/journal.pone.0275623. PMID: 35075458 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

