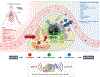
Mitochondrial signal transduction
- PMID: 36323233
- PMCID: PMC9692202
- DOI: 10.1016/j.cmet.2022.10.008
Mitochondrial signal transduction
Abstract
The analogy of mitochondria as powerhouses has expired. Mitochondria are living, dynamic, maternally inherited, energy-transforming, biosynthetic, and signaling organelles that actively transduce biological information. We argue that mitochondria are the processor of the cell, and together with the nucleus and other organelles they constitute the mitochondrial information processing system (MIPS). In a three-step process, mitochondria (1) sense and respond to both endogenous and environmental inputs through morphological and functional remodeling; (2) integrate information through dynamic, network-based physical interactions and diffusion mechanisms; and (3) produce output signals that tune the functions of other organelles and systemically regulate physiology. This input-to-output transformation allows mitochondria to transduce metabolic, biochemical, neuroendocrine, and other local or systemic signals that enhance organismal adaptation. An explicit focus on mitochondrial signal transduction emphasizes the role of communication in mitochondrial biology. This framework also opens new avenues to understand how mitochondria mediate inter-organ processes underlying human health.
Keywords: amplification; communication; energy; evolution; health; membrane potential; metabokines; mito-nuclear signaling; mitochondrial networks; mitokines; mitotypes; receptors; signal transduction; steroid hormones; stress responses; tissue-specific.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests O.S.S. is a co-founder of Capacity and Inspire Bio.
Figures





References
-
- Altman R (1890). Die Elementarorganismen und ihre Beziehungen zu den Zellen (Veit).
-
- Siekevitz P. (1957). Powerhouse of the Cell (Scientific American). https://www.scientificamerican.com/article/powerhouse-of-the-cell/.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

