Disruption of mTORC1 rescues neuronal overgrowth and synapse function dysregulated by Pten loss
- PMID: 36323257
- PMCID: PMC9743803
- DOI: 10.1016/j.celrep.2022.111574
Disruption of mTORC1 rescues neuronal overgrowth and synapse function dysregulated by Pten loss
Abstract
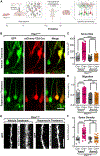
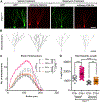
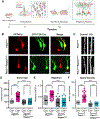
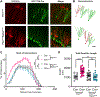
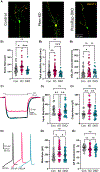
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is a negative regulator of AKT/mTOR signaling pathway. Mutations in PTEN are found in patients with autism, epilepsy, or macrocephaly. In mouse models, Pten loss results in neuronal hypertrophy, hyperexcitability, seizures, and ASD-like behaviors. The underlying molecular mechanisms of these phenotypes are not well delineated. We determined which of the Pten loss-driven aberrations in neuronal form and function are orchestrated by downstream mTOR complex 1 (mTORC1). Rapamycin-mediated inhibition of mTORC1 prevented increase in soma size, migration, spine density, and dendritic overgrowth in Pten knockout dentate gyrus granule neurons. Genetic knockout of Raptor to disrupt mTORC1 complex formation blocked Pten loss-mediated neuronal hypertrophy. Electrophysiological recordings revealed that genetic disruption of mTORC1 rescued Pten loss-mediated increase in excitatory synaptic transmission. We have identified an essential role for mTORC1 in orchestrating Pten loss-driven neuronal hypertrophy and synapse formation.
Keywords: CP: Cell biology; CP: Neuroscience; PTEN; Raptor; autism; dendrite; mTOR; rapamycin; synapse.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Authors declare no competing interests.
Figures


References
-
- Cardamone M, Flanagan D, Mowat D, Kennedy SE, Chopra M, and Lawson JA (2014). Mammalian target of rapamycin inhibitors for intractable epilepsy and subependymal giant cell astrocytomas in tuberous sclerosis complex. J. Pediatr 164, 1195–1200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

