HIF1α-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex
- PMID: 36323673
- PMCID: PMC9630315
- DOI: 10.1038/s41467-022-34252-2
HIF1α-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex
Abstract
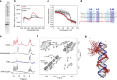
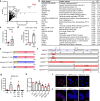
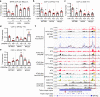
DNA:DNA:RNA triplexes that are formed through Hoogsteen base-pairing of the RNA in the major groove of the DNA duplex have been observed in vitro, but the extent to which these interactions occur in cells and how they impact cellular functions remains elusive. Using a combination of bioinformatic techniques, RNA/DNA pulldown and biophysical studies, we set out to identify functionally important DNA:DNA:RNA triplex-forming long non-coding RNAs (lncRNA) in human endothelial cells. The lncRNA HIF1α-AS1 was retrieved as a top hit. Endogenous HIF1α-AS1 reduces the expression of numerous genes, including EPH Receptor A2 and Adrenomedullin through DNA:DNA:RNA triplex formation by acting as an adapter for the repressive human silencing hub complex (HUSH). Moreover, the oxygen-sensitive HIF1α-AS1 is down-regulated in pulmonary hypertension and loss-of-function approaches not only result in gene de-repression but also enhance angiogenic capacity. As exemplified here with HIF1α-AS1, DNA:DNA:RNA triplex formation is a functionally important mechanism of trans-acting gene expression control.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. doi: 10.1021/ja01565a074. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

