Deep learning empowered volume delineation of whole-body organs-at-risk for accelerated radiotherapy
- PMID: 36323677
- PMCID: PMC9630370
- DOI: 10.1038/s41467-022-34257-x
Deep learning empowered volume delineation of whole-body organs-at-risk for accelerated radiotherapy
Abstract
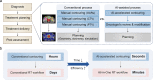
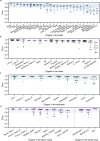
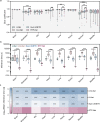
In radiotherapy for cancer patients, an indispensable process is to delineate organs-at-risk (OARs) and tumors. However, it is the most time-consuming step as manual delineation is always required from radiation oncologists. Herein, we propose a lightweight deep learning framework for radiotherapy treatment planning (RTP), named RTP-Net, to promote an automatic, rapid, and precise initialization of whole-body OARs and tumors. Briefly, the framework implements a cascade coarse-to-fine segmentation, with adaptive module for both small and large organs, and attention mechanisms for organs and boundaries. Our experiments show three merits: 1) Extensively evaluates on 67 delineation tasks on a large-scale dataset of 28,581 cases; 2) Demonstrates comparable or superior accuracy with an average Dice of 0.95; 3) Achieves near real-time delineation in most tasks with <2 s. This framework could be utilized to accelerate the contouring process in the All-in-One radiotherapy scheme, and thus greatly shorten the turnaround time of patients.
© 2022. The Author(s).
Conflict of interest statement
F.S., J.W., M.H., Q.Z., Y.W., Y.S., Y.C., Y.Y., X.C., Y.Z., X.S.Z., Y.G., and D.S. are employees of Shanghai United Imaging Intelligence Co., Ltd.; W.Z., and J.Z. are employees of Shanghai United Imaging Healthcare Co., Ltd. The companies have no role in designing and performing the surveillance and analyzing and interpreting the data. All other authors report no conflicts of interest relevant to this article.
Figures




References
-
- Sung H, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA-Cancer J. Clin. 2021;71:7–33. - PubMed
-
- Wei W, et al. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol. 2020;21:e342–e349. - PubMed
-
- Atun R, et al. Expanding global access to radiotherapy. Lancet Oncol. 2015;16:1153–1186. - PubMed
-
- Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

