FOXA1 repression drives lineage plasticity and immune heterogeneity in bladder cancers with squamous differentiation
- PMID: 36323682
- PMCID: PMC9630410
- DOI: 10.1038/s41467-022-34251-3
FOXA1 repression drives lineage plasticity and immune heterogeneity in bladder cancers with squamous differentiation
Erratum in
-
Author Correction: FOXA1 repression drives lineage plasticity and immune heterogeneity in bladder cancers with squamous differentiation.Nat Commun. 2022 Dec 23;13(1):7920. doi: 10.1038/s41467-022-35644-0. Nat Commun. 2022. PMID: 36564410 Free PMC article. No abstract available.
Abstract
Cancers arising from the bladder urothelium often exhibit lineage plasticity with regions of urothelial carcinoma adjacent to or admixed with regions of divergent histomorphology, most commonly squamous differentiation. To define the biologic basis for and clinical significance of this morphologic heterogeneity, here we perform integrated genomic analyses of mixed histology bladder cancers with separable regions of urothelial and squamous differentiation. We find that squamous differentiation is a marker of intratumoral genomic and immunologic heterogeneity in patients with bladder cancer and a biomarker of intrinsic immunotherapy resistance. Phylogenetic analysis confirms that in all cases the urothelial and squamous regions are derived from a common shared precursor. Despite the presence of marked genomic heterogeneity between co-existent urothelial and squamous differentiated regions, no recurrent genomic alteration exclusive to the urothelial or squamous morphologies is identified. Rather, lineage plasticity in bladder cancers with squamous differentiation is associated with loss of expression of FOXA1, GATA3, and PPARG, transcription factors critical for maintenance of urothelial cell identity. Of clinical significance, lineage plasticity and PD-L1 expression is coordinately dysregulated via FOXA1, with patients exhibiting morphologic heterogeneity pre-treatment significantly less likely to respond to immune checkpoint inhibitors.
© 2022. The Author(s).
Conflict of interest statement
E.J.P. received honorarium from UpToDate, and received research funding from and is on the scientific advisory boards for Janssen Pharmaceuticals, Merck & Co. Inc., QED Therapeutics, UroGen Pharma and Steba Biotech. D.B.S. has consulted for/received honoraria from Pfizer, Loxo/Lilly Oncology, FORE Therapeutics, Vividion Therapeutics, Scorpion Therapeutics, Fog Pharma, and BridgeBio. D.J.D. received a research grant from Bristol-Myers-Squibb. H.A.A has consulted for Bristol-Myers-Squibb, AstraZeneca, Janssen Biotech, and Paige.ai. M.J. received research grant from AstraZeneca, Pfizer, and Eisai (drug only to Institution), and is on the advisory board for Seagen. J.S. is a consultant for Janssen Research & Development, LLC. S.A.F. received research support from AstraZeneca, Genentech/Roche, is a consultant/advisory board member for Merck and BioNTech, and owns stock/equity interest in Urogen, Allogene Therapeutics, Neogene Therapeutics, Kronos Bio, ByHeart, 76Bio, Vida Ventures, Inconovir, and Doximity. B.H.B is a consultant to Olympus corporation. D.F.B. reports personal fees from Bristol Myers Squibb and Merck; consulting/advisory role for Merck, Dragonfly Therapeutics, Fidia Farmaceutici S.p.A., and Bristol Myers Squibb Foundation; Travel/accommodations/expenses from Merck; and institutional research funding from Novartis, Merck, Bristol-Myers Squibb, AstraZeneca, Astellas Pharma, and Seattle Genetics/Astellas. Y.C. reports stock ownership in Oric Pharmaceuticals and sponsored Research from Foghorn Therapeutics. J.E.R is a Consultant for Seagen, Astellas, Bayer, AstraZeneca, QED Therapeutics, Merck, Genentech, Infinity, Gilead, Boehringer Ingelheim, Tyra, Mirati, Pfizer, EMD-Serono; reports sponsored research from Seagen, Astellas, Bayer, AstraZeneca, QED Therapeutics, Genentech; and received honoraria from Pfizer and EMD-Serono. The remaining authors declare no conflicts of interest.
Figures
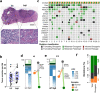






References
-
- De Santis M, et al. Randomized phase II/III trial assessing gemcitabine/carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer who are unfit for cisplatin-based chemotherapy: EORTC study 30986. J. Clin. Oncol. 2012;30:191–199. doi: 10.1200/JCO.2011.37.3571. - DOI - PMC - PubMed
-
- Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

