Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells
- PMID: 36323770
- PMCID: PMC9628577
- DOI: 10.1038/s41598-022-21034-5
Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells
Abstract
SARS coronavirus 2 (SARS-CoV-2) has caused an ongoing global pandemic with significant mortality and morbidity. At this time, the only FDA-approved therapeutic for COVID-19 is remdesivir, a broad-spectrum antiviral nucleoside analog. Efficacy is only moderate, and improved treatment strategies are urgently needed. To accomplish this goal, we devised a strategy to identify compounds that act synergistically with remdesivir in preventing SARS-CoV-2 replication. We conducted combinatorial high-throughput screening in the presence of submaximal remdesivir concentrations, using a human lung epithelial cell line infected with a clinical isolate of SARS-CoV-2. This identified 20 approved drugs that act synergistically with remdesivir, many with favorable pharmacokinetic and safety profiles. Strongest effects were observed with established antivirals, Hepatitis C virus nonstructural protein 5A (HCV NS5A) inhibitors velpatasvir and elbasvir. Combination with their partner drugs sofosbuvir and grazoprevir further increased efficacy, increasing remdesivir's apparent potency > 25-fold. We report that HCV NS5A inhibitors act on the SARS-CoV-2 exonuclease proofreader, providing a possible explanation for the synergy observed with nucleoside analog remdesivir. FDA-approved Hepatitis C therapeutics Epclusa® (velpatasvir/sofosbuvir) and Zepatier® (elbasvir/grazoprevir) could be further optimized to achieve potency and pharmacokinetic properties that support clinical evaluation in combination with remdesivir.
© 2022. The Author(s).
Conflict of interest statement
JS is inventor in a patent application on combination treatments for SARS-CoV-2, owned by the University of California. U.S. Patent Application Serial No. 63/053,208, entitled “COMPOSITIONS AND METHODS FOR TREATING VIRAL INFECTIONS” relates to aspects of this work and was filed on July 17th, 2020. XHW, EW, EVD, SBB, LY, CZ, JSt, CDD, TG, XW, SJ, CT, MC, WX, DP, CM, AG, TT, JJR, JJ, AN, SS declare no potential conflict of interest.
Figures
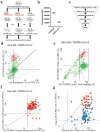
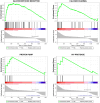
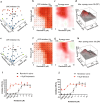
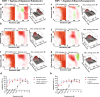
Similar articles
-
Remdesivir for the treatment of COVID-19.Cochrane Database Syst Rev. 2023 Jan 25;1(1):CD014962. doi: 10.1002/14651858.CD014962.pub2. Cochrane Database Syst Rev. 2023. PMID: 36695483 Free PMC article.
-
Clinical efficacy of antiviral agents against coronavirus disease 2019: A systematic review of randomized controlled trials.J Microbiol Immunol Infect. 2021 Oct;54(5):767-775. doi: 10.1016/j.jmii.2021.05.011. Epub 2021 Jun 26. J Microbiol Immunol Infect. 2021. PMID: 34253490 Free PMC article.
-
Direct-acting antivirals for chronic hepatitis C.Cochrane Database Syst Rev. 2017 Sep 18;9(9):CD012143. doi: 10.1002/14651858.CD012143.pub3. Cochrane Database Syst Rev. 2017. PMID: 28922704 Free PMC article.
-
A mutation in the coronavirus nsp13-helicase impairs enzymatic activity and confers partial remdesivir resistance.mBio. 2023 Aug 31;14(4):e0106023. doi: 10.1128/mbio.01060-23. Epub 2023 Jun 20. mBio. 2023. PMID: 37338298 Free PMC article.
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
Cited by
-
Efficacy and safety of the sofosbuvir/velpatasvir combination for the treatment of patients with early mild to moderate COVID-19.Sci Rep. 2022 Apr 6;12(1):5771. doi: 10.1038/s41598-022-09741-5. Sci Rep. 2022. PMID: 35388092 Free PMC article.
-
SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA.PLoS Biol. 2021 Oct 11;19(10):e3001425. doi: 10.1371/journal.pbio.3001425. eCollection 2021 Oct. PLoS Biol. 2021. PMID: 34634033 Free PMC article.
-
Drug Combinations as a First Line of Defense against Coronaviruses and Other Emerging Viruses.mBio. 2021 Dec 21;12(6):e0334721. doi: 10.1128/mbio.03347-21. Epub 2021 Dec 21. mBio. 2021. PMID: 34933447 Free PMC article. Review.
-
SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA.bioRxiv [Preprint]. 2021 Mar 29:2020.09.14.295824. doi: 10.1101/2020.09.14.295824. bioRxiv. 2021. Update in: PLoS Biol. 2021 Oct 11;19(10):e3001425. doi: 10.1371/journal.pbio.3001425. PMID: 32995779 Free PMC article. Updated. Preprint.
-
In Vitro Combinatorial Activity of Direct Acting Antivirals and Monoclonal Antibodies against the Ancestral B.1 and BQ.1.1 SARS-CoV-2 Viral Variants.Viruses. 2024 Jan 23;16(2):168. doi: 10.3390/v16020168. Viruses. 2024. PMID: 38399944 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

