Regulatory mechanism of fibrosis-related genes in patients with heart failure
- PMID: 36324504
- PMCID: PMC9618712
- DOI: 10.3389/fgene.2022.1032572
Regulatory mechanism of fibrosis-related genes in patients with heart failure
Abstract
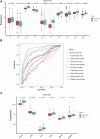
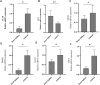
Background: Heart failure (HF) is a complex clinical syndrome characterized by the inability to match cardiac output with metabolic needs. Research on regulatory mechanism of fibrosis-related genes in patients with HF is very limited. In order to understand the mechanism of fibrosis in the development and progression of HF, fibrosis -related hub genes in HF are screened and verified. Methods: RNA sequencing data was obtained from the Gene Expression Omnibus (GEO) cohorts to identify differentially expressed genes (DEGs). Thereafter, fibrosis-related genes were obtained from the GSEA database and that associated with HF were screened out. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis was carried out to analyze the biological function of fibrosis-related DEGs. The protein-protein interaction (PPI) network of hub genes was constructed via the STRING database. Moreover, the diagnostic value of hub genes for HF was confirmed using ROC curves and expression analysis. Finally, quantitative real time PCR was used to detect the expression levels of mRNAs. Results: A total of 3, 469 DEGs were identified closely related to HF, and 1, 187 fibrosis-related DEGs were obtained and analyzed for GO and KEGG enrichment. The enrichment results of fibrosis-related DEGs were consistent with that of DEGs. A total of 10 hub genes (PPARG, KRAS, JUN, IL10, TLR4, STAT3, CXCL8, CCL2, IL6, IL1β) were selected via the PPI network. Receiver operating characteristic curve analysis was estimated in the test cohort, and 6 genes (PPARG, KRAS, JUN, IL10, TLR4, STAT3) with AUC more than 0.7 were identified as diagnosis genes. Moreover, miRNA-mRNA and TF-mRNA regulatory networks were constructed. Finally, quantitative real time PCR revealed these 6 genes may be used as the potential diagnostic biomarkers of HF. Conclusion: In this study, 10 fibrosis-related hub genes in the HF were identified and 6 of them were demonstrated as potential diagnostic biomarkers for HF.
Keywords: cardiac remodeling; diagnosis; fibrosis; heart failure; hub gene.
Copyright © 2022 Tao, Gao, Qian, Cao, Han and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Integrative analysis of potential biomarkers and immune cell infiltration in Parkinson's disease.Brain Res Bull. 2021 Dec;177:53-63. doi: 10.1016/j.brainresbull.2021.09.010. Epub 2021 Sep 16. Brain Res Bull. 2021. PMID: 34536521
-
Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis.BMC Cardiovasc Disord. 2021 Jul 4;21(1):329. doi: 10.1186/s12872-021-02146-8. BMC Cardiovasc Disord. 2021. PMID: 34218797 Free PMC article.
-
Exploration of the pathogenesis of Sjögren's syndrome via DNA methylation and transcriptome analyses.Clin Rheumatol. 2022 Sep;41(9):2765-2777. doi: 10.1007/s10067-022-06200-4. Epub 2022 May 13. Clin Rheumatol. 2022. PMID: 35562622
-
STAT4 and COL1A2 are potential diagnostic biomarkers and therapeutic targets for heart failure comorbided with depression.Brain Res Bull. 2022 Jun 15;184:68-75. doi: 10.1016/j.brainresbull.2022.03.014. Epub 2022 Mar 31. Brain Res Bull. 2022. PMID: 35367598 Review.
-
Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis.Medicine (Baltimore). 2020 Oct 16;99(42):e22434. doi: 10.1097/MD.0000000000022434. Medicine (Baltimore). 2020. PMID: 33080677 Free PMC article.
Cited by
-
A Meta-Analysis Approach to Gene Regulatory Network Inference Identifies Key Regulators of Cardiovascular Diseases.Int J Mol Sci. 2024 Apr 11;25(8):4224. doi: 10.3390/ijms25084224. Int J Mol Sci. 2024. PMID: 38673810 Free PMC article.
-
Novel Impact of Colchicine on Interleukin-10 Expression in Acute Myocardial Infarction: An Integrative Approach.J Clin Med. 2024 Aug 7;13(16):4619. doi: 10.3390/jcm13164619. J Clin Med. 2024. PMID: 39200761 Free PMC article.
References
-
- Boye P., Abdel-Aty H., Zacharzowsky U., Bohl S., Schwenke C., van der Geest R. J., et al. (2011). Prediction of life-threatening arrhythmic events in patients with chronic myocardial infarction by contrast-enhanced CMR. JACC. Cardiovasc. Imaging 4 (8), 871–879. 10.1016/j.jcmg.2011.04.014 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

