Identification of genes modified by N6-methyladenosine in patients with colorectal cancer recurrence
- PMID: 36324506
- PMCID: PMC9618605
- DOI: 10.3389/fgene.2022.1043297
Identification of genes modified by N6-methyladenosine in patients with colorectal cancer recurrence
Abstract
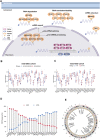
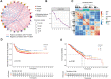
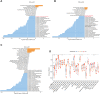
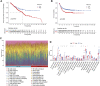
Background: Recent studies demonstrate that N6-methyladenosine (m6A) methylation plays a crucial role in colorectal cancer (CRC). Therefore, we conducted a comprehensive analysis to assess the m6A modification patterns and identify m6A-modified genes in patients with CRC recurrence. Methods: The m6A modification patterns were comprehensively evaluated by the NMF algorithm based on the levels of 27 m6A regulators, and tumor microenvironment (TME) cell-infiltrating characteristics of these modification patterns were systematically assessed by ssGSEA and CIBERSORT algorithms. The principal component analysis algorithm based on the m6A scoring scheme was used to explore the m6A modification patterns of individual tumors with immune responses. The weighted correlation network analysis and univariable and multivariable Cox regression analyses were applied to identify m6A-modified gene signatures. The single-cell expression dataset of CRC samples was used to explore the tumor microenvironment affected by these signatures. Results: Three distinct m6A modification patterns with significant recurrence-free survival (RFS) were identified in 804 CRC patients. The TME characterization revealed that the m6A modification pattern with longer RFS exhibited robust immune responses. CRC patients were divided into high- and low-score subgroups according to the m6A score individually, which was obtained from the m6A-related signature genes. The patients with low m6A scores had both longer RFS and overall survival (OS) with altered immune cell infiltration. Notably, m6A-modified genes showed significant differences related to the prognosis of CRC patients in the meta-GEO cohort and TCGA cohort. Single-cell expression indicated that ALVRL1 was centrally distributed in endothelial tip cells and stromal cells. Conclusion: The m6A modification plays an indispensable role in the formation of TME diversity and complexity. Importantly, the signatures (TOP2A, LRRC58, HAUS6, SMC4, ACVRL1, and KPNB1) were identified as m6A-modified genes associated with CRC recurrence, thereby serving as a promising predictive biomarker or therapeutic target for patients with CRC recurrence.
Keywords: colorectal cancer; m6A methylation modification; overall survival; recurrence; tumor immune microenvironment.
Copyright © 2022 Zhu, Huang, Yu, Shou, Zhang, Xie, Liang, Sun, Feng, Duan, Zhang, Xiang, Sui, Jin, Yu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The significance of m6A RNA methylation modification in prognosis and tumor microenvironment immune infiltration of cervical cancer.Medicine (Baltimore). 2022 Jul 1;101(26):e29818. doi: 10.1097/MD.0000000000029818. Medicine (Baltimore). 2022. PMID: 35777046 Free PMC article.
-
m6A regulator-based methylation modification patterns characterized by distinct tumor microenvironment immune profiles in colon cancer.Theranostics. 2021 Jan 1;11(5):2201-2217. doi: 10.7150/thno.52717. eCollection 2021. Theranostics. 2021. PMID: 33500720 Free PMC article.
-
N6-Methyladenosine Modification Patterns and Tumor Microenvironment Immune Characteristics Associated With Clinical Prognosis Analysis in Stomach Adenocarcinoma.Front Cell Dev Biol. 2022 Jun 15;10:913307. doi: 10.3389/fcell.2022.913307. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35813200 Free PMC article.
-
Interaction between N6-methyladenosine modification and the tumor microenvironment in colorectal cancer.Mol Med. 2023 Sep 22;29(1):129. doi: 10.1186/s10020-023-00726-2. Mol Med. 2023. PMID: 37737134 Free PMC article. Review.
-
N6-methyladenosine (m6A) RNA modification in tumor immunity.Cancer Biol Med. 2022 Mar 8;19(4):385-97. doi: 10.20892/j.issn.2095-3941.2021.0534. Online ahead of print. Cancer Biol Med. 2022. PMID: 35254013 Free PMC article. Review.
Cited by
-
A genome-wide SNP-SNP interaction analysis exploring novel interacting loci associated with the risk of recurrence in colorectal cancer.PLoS One. 2025 Jun 18;20(6):e0321967. doi: 10.1371/journal.pone.0321967. eCollection 2025. PLoS One. 2025. PMID: 40531904 Free PMC article.
-
FoxO1 promotes ovarian cancer by increasing transcription and METTL14-mediated m6A modification of SMC4.Cancer Sci. 2024 Apr;115(4):1224-1240. doi: 10.1111/cas.16120. Epub 2024 Feb 25. Cancer Sci. 2024. PMID: 38403332 Free PMC article.
-
A GWAS Suggesting Genetic Modifiers to Increase the Risk of Colorectal Cancer from Antibiotic Use.Cancers (Basel). 2024 Dec 24;17(1):12. doi: 10.3390/cancers17010012. Cancers (Basel). 2024. PMID: 39796643 Free PMC article.
References
-
- Allen W. L., Dunne P. D., McDade S., Scanlon E., Loughrey M., Coleman H., et al. (2018). Transcriptional subtyping and CD8 immunohistochemistry identifies poor prognosis stage II/III colorectal cancer patients who benefit from adjuvant chemotherapy. JCO Precis. Oncol. 17, 00241. 10.1200/PO.17.00241 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

