This is a preprint.
Modulation of type I interferon responses potently inhibits SARS-CoV-2 replication and inflammation in rhesus macaques
- PMID: 36324810
- PMCID: PMC9628196
- DOI: 10.1101/2022.10.21.512606
Modulation of type I interferon responses potently inhibits SARS-CoV-2 replication and inflammation in rhesus macaques
Update in
-
Modulation of type I interferon responses potently inhibits SARS-CoV-2 replication and inflammation in rhesus macaques.Sci Immunol. 2023 Jul 28;8(85):eadg0033. doi: 10.1126/sciimmunol.adg0033. Epub 2023 Jul 28. Sci Immunol. 2023. PMID: 37506197 Free PMC article.
Abstract
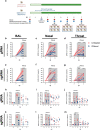
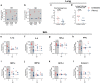
Type-I interferons (IFN-I) are critical mediators of innate control of viral infections, but also drive recruitment of inflammatory cells to sites of infection, a key feature of severe COVID-19. Here, and for the first time, IFN-I signaling was modulated in rhesus macaques (RMs) prior to and during acute SARS-CoV-2 infection using a mutated IFNα2 (IFN-modulator; IFNmod), which has previously been shown to reduce the binding and signaling of endogenous IFN-I. In SARS-CoV-2-infected RMs, IFNmod reduced both antiviral and inflammatory ISGs. Notably, IFNmod treatment resulted in a potent reduction in (i) SARS-CoV-2 viral load in Bronchoalveolar lavage (BAL), upper airways, lung, and hilar lymph nodes; (ii) inflammatory cytokines, chemokines, and CD163+MRC1-inflammatory macrophages in BAL; and (iii) expression of Siglec-1, which enhances SARS-CoV-2 infection and predicts disease severity, on circulating monocytes. In the lung, IFNmod also reduced pathogenesis and attenuated pathways of inflammasome activation and stress response during acute SARS-CoV-2 infection. This study, using an intervention targeting both IFN-α and IFN-β pathways, shows that excessive inflammation driven by type 1 IFN critically contributes to SARS-CoV-2 pathogenesis in RMs, and demonstrates the potential of IFNmod to limit viral replication, SARS-CoV-2 induced inflammation, and COVID-19 severity.
Conflict of interest statement
Competing Interests
The authors have nothing to disclose.
Figures




References
-
- Isaacs A. & Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147, 258–267 (1957). - PubMed
-
- Isaacs A., Lindenmann J. & Valentine R.C. Virus interference. II. Some properties of interferon. Proc R Soc Lond B Biol Sci 147, 268–273 (1957). - PubMed
-
- Schoggins J.W. Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol 6, 567–584 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
