SPREAD 4: online visualisation of pathogen phylogeographic reconstructions
- PMID: 36325034
- PMCID: PMC9615431
- DOI: 10.1093/ve/veac088
SPREAD 4: online visualisation of pathogen phylogeographic reconstructions
Abstract
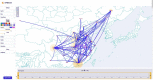
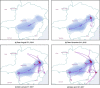
Phylogeographic analyses aim to extract information about pathogen spread from genomic data, and visualising spatio-temporal reconstructions is a key aspect of this process. Here we present SPREAD 4, a feature-rich web-based application that visualises estimates of pathogen dispersal resulting from Bayesian phylogeographic inference using BEAST on a geographic map, offering zoom-and-filter functionality and smooth animation over time. SPREAD 4 takes as input phylogenies with both discrete and continuous location annotation and offers customised visualisation as well as generation of publication-ready figures. SPREAD 4 now features account-based storage and easy sharing of visualisations by means of unique web addresses. SPREAD 4 is intuitive to use and is available online at https://spreadviz.org, with an accompanying web page containing answers to frequently asked questions at https://beast.community/spread4.
Keywords: BEAST; Bayesian inference; phylogeography; viral spread; visualisation.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Aksamentov I. et al. (2021) ‘Nextclade: clade assignment, mutation calling and quality control for viral genomes’, Journal of open source software, 6: 3773. doi: 10.21105/joss.03773 - DOI
-
- Bernasconi A. and Grandi S. (2021) ‘A conceptual model for geo-online exploratory data visualization: The case of the COVID-19 pandemic’, Information, 12: 69. doi: 10.3390/info12020069 - DOI

