The His-tag as a decoy modulating preferred orientation in cryoEM
- PMID: 36325274
- PMCID: PMC9619061
- DOI: 10.3389/fmolb.2022.912072
The His-tag as a decoy modulating preferred orientation in cryoEM
Abstract
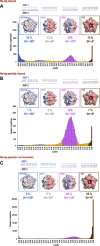
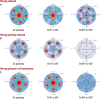
The His-tag is a widely used affinity tag that facilitates purification by means of affinity chromatography of recombinant proteins for functional and structural studies. We show here that His-tag presence affects how coproheme decarboxylase interacts with the air-water interface during grid preparation for cryoEM. Depending on His-tag presence or absence, we observe significant changes in patterns of preferred orientation. Our analysis of particle orientations suggests that His-tag presence can mask the hydrophobic and hydrophilic patches on a protein's surface that mediate the interactions with the air-water interface, while the hydrophobic linker between a His-tag and the coding sequence of the protein may enhance other interactions with the air-water interface. Our observations suggest that tagging, including rational design of the linkers between an affinity tag and a protein of interest, offer a promising approach to modulating interactions with the air-water interface.
Keywords: AWI; His-tag; air-water interface; cryo-electron microscopy single particle reconstruction; cryoEM SPR; preferred orientation; visualization.
Copyright © 2022 Bromberg, Cai, Guo, Plymire, Emde, Puzio, Borek and Otwinowski.
Conflict of interest statement
RB, YG, DB, and ZO are co-founders of Ligo Analytics, a company that develops software for cryogenic electron microscopy. YG serves as the CEO of Ligo Analytics, RB is currently employed by Ligo Analytics, while DP was employed by Ligo Analytics and UT Southwestern in the past. ZO is a co-founder of HKL Research, a company that develops and distributes software for X-ray crystallography. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Barth M., Bryan R. K., Hegerl R. (1989). Approximation of missing-cone data in 3d electron-microscopy. Ultramicroscopy 31 (4), 365–378. 10.1016/0304-3991(89)90335-5 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

