HLA-C dysregulation as a possible mechanism of immune evasion in SARS-CoV-2 and other RNA-virus infections
- PMID: 36325330
- PMCID: PMC9618630
- DOI: 10.3389/fimmu.2022.1011829
HLA-C dysregulation as a possible mechanism of immune evasion in SARS-CoV-2 and other RNA-virus infections
Abstract
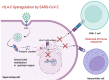
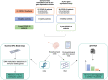
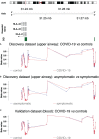
One of the mechanisms by which viruses can evade the host's immune system is to modify the host's DNA methylation pattern. This work aims to investigate the DNA methylation and gene expression profile of COVID-19 patients, divided into symptomatic and asymptomatic, and healthy controls, focusing on genes involved in the immune response. In this study, changes in the methylome of COVID-19 patients' upper airways cells, the first barrier against respiratory infections and the first cells presenting viral antigens, are shown for the first time. Our results showed alterations in the methylation pattern of genes encoding proteins implicated in the response against pathogens, in particular the HLA-C gene, also important for the T-cell mediated memory response. HLA-C expression significantly decreases in COVID-19 patients, especially in those with a more severe prognosis and without other possibly confounding co-morbidities. Moreover, our bionformatic analysis revealed that the identified methylation alteration overlaps with enhancers regulating HLA-C expression, suggesting an additional mechanism exploited by SARS-CoV-2 to inhibit this fundamental player in the host's immune response. HLA-C could therefore represent both a prognostic marker and an excellent therapeutic target, also suggesting a preventive intervention that conjugate a virus-specific antigenic stimulation with an adjuvant increasing the T-cell mediated memory response.
Keywords: COVID-19; DNA methylation; HLA-C; SARS-CoV-2; enhancer transcriptional regulation; gene expression; symptomatic and asymptomatic COVID-19; upper airways cells.
Copyright © 2022 Loi, Moi, Cabras, Arduino, Costanzo, Del Giacco, Erlich, Firinu, Caddori and Zavattari.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

