Genomic comparisons confirm Giardia duodenalis sub-assemblage AII as a unique species
- PMID: 36325462
- PMCID: PMC9618722
- DOI: 10.3389/fcimb.2022.1010244
Genomic comparisons confirm Giardia duodenalis sub-assemblage AII as a unique species
Abstract
Giardia duodenalis is a parasitic flagellated protozoan which infects a wide range of mammalian hosts, including humans, and is subdivided into at least eight genetic assemblages commonly thought to represent cryptic species. Molecular studies have shown that G. duodenalis assemblage A, which parasitizes humans and animals, contains several phylogenetically distinct groupings known as sub-assemblages. Molecular studies employing poor phylogenetic-resolution markers routinely recover these sub-assemblages, implying that they represent evolutionarily distinct clades and possibly cryptic species, a hypothesis which is supported by epidemiologic trends. Here, we further tested this hypothesis by using available data from 41 whole genomes to characterize sub-assemblages and coalescent techniques for statistical estimation of species boundaries coupled to functional gene content analysis, thereby assessing the stability and distinctiveness of clades. Our analysis revealed two new sub-assemblage clades as well as novel signatures of gene content geared toward differential host adaptation and population structuring via vertical inheritance rather than recombination or panmixia. We formally propose sub-assemblage AII as a new species, Giardia hominis, while preserving the name Giardia duodenalis for sub-assemblage AI. Additionally, our bioinformatic methods broadly address the challenges of identifying cryptic microbial species to advance our understanding of emerging disease epidemiology, which should be broadly applicable to other lower eukaryotic taxa of interest. Giardia hominis n. sp. Zoobank LSID: urn:lsid: zoobank.org:pub:4298F3E1-E3EF-4977-B9DD-5CC59378C80E.
Keywords: Giardia; assemblage A; comparative genomics; cryptic species; phylogenetics; zoonotic diseases.
Copyright © 2022 Seabolt, Roellig and Konstantinidis.
Conflict of interest statement
Author MS was employed by Leidos Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
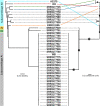
Figures




Similar articles
-
Proposed taxonomic revision of Giardia duodenalis.Infect Genet Evol. 2023 Jul;111:105430. doi: 10.1016/j.meegid.2023.105430. Epub 2023 Mar 25. Infect Genet Evol. 2023. PMID: 36972861
-
Prevalence of Giardia duodenalis assemblages and sub-assemblages in symptomatic patients from Damascus city and its suburbs.Infect Genet Evol. 2017 Jan;47:155-160. doi: 10.1016/j.meegid.2016.11.030. Epub 2016 Dec 3. Infect Genet Evol. 2017. PMID: 27919804
-
Determination of Giardia duodenalis assemblages and multi-locus genotypes in patients with sporadic giardiasis from England.Parasit Vectors. 2015 Sep 4;8:444. doi: 10.1186/s13071-015-1059-z. Parasit Vectors. 2015. PMID: 26338670 Free PMC article.
-
Giardia duodenalis genetic assemblages and hosts.Parasite. 2016;23:13. doi: 10.1051/parasite/2016013. Epub 2016 Mar 16. Parasite. 2016. PMID: 26984116 Free PMC article. Review.
-
A review of the molecular epidemiology of Cryptosporidium spp. and Giardia duodenalis in the Middle East and North Africa (MENA) region.Infect Genet Evol. 2022 Mar;98:105212. doi: 10.1016/j.meegid.2022.105212. Epub 2022 Jan 20. Infect Genet Evol. 2022. PMID: 35065302 Review.
Cited by
-
Genomic survey maps differences in the molecular complement of vesicle formation machinery between Giardia intestinalis assemblages.PLoS Negl Trop Dis. 2023 Dec 18;17(12):e0011837. doi: 10.1371/journal.pntd.0011837. eCollection 2023 Dec. PLoS Negl Trop Dis. 2023. PMID: 38109380 Free PMC article.
-
Comprehensive analysis of flavohemoprotein copy number variation in Giardia intestinalis: exploring links to metronidazole resistance.Parasit Vectors. 2024 Aug 10;17(1):336. doi: 10.1186/s13071-024-06392-5. Parasit Vectors. 2024. PMID: 39127700 Free PMC article.
-
Asymptomatic carriage of intestinal protists is common in children in Lusaka Province, Zambia.PLoS Negl Trop Dis. 2024 Dec 13;18(12):e0012717. doi: 10.1371/journal.pntd.0012717. eCollection 2024 Dec. PLoS Negl Trop Dis. 2024. PMID: 39671429 Free PMC article.
-
Molecular characterization of common zoonotic protozoan parasites and bacteria causing diarrhea in dairy calves in Ningxia Hui Autonomous Region, China.Parasite. 2024;31:60. doi: 10.1051/parasite/2024059. Epub 2024 Oct 1. Parasite. 2024. PMID: 39353100 Free PMC article.
-
Assemblage characterization of Giardia duodenalis in South Khorasan province, eastern Iran, using HRM real-time PCR method.Mol Biol Rep. 2024 Jan 18;51(1):127. doi: 10.1007/s11033-023-09001-3. Mol Biol Rep. 2024. PMID: 38236550
References
-
- Adam R. D., Dahlstrom E. W., Martens C. A., Bruno D. P., Barbian K. D., Ricklefs S. M., et al. (2013). Genome sequencing of giardia lamblia genotypes A2 and b isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and e (WB and pig). Genome Biol. Evol. 5, 2498–2511. doi: 10.1093/gbe/evt197 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

