Boroleucine-Derived Covalent Inhibitors of the ZIKV Protease
- PMID: 36325810
- PMCID: PMC10100045
- DOI: 10.1002/cmdc.202200336
Boroleucine-Derived Covalent Inhibitors of the ZIKV Protease
Abstract
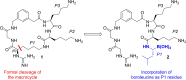
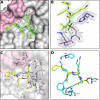
The Zika virus (ZIKV) remains a potential threat to the public health due to the lack of both an approved vaccination or a specific treatment. In this work, a series of peptidic inhibitors of the ZIKV protease with boroleucine as P1 residue was synthesized. The highest affinities with Ki values down to 8 nM were observed for compounds with basic residues in both P2 and P3 position and at the N-terminus. The low potency of reference compounds containing leucine, leucine-amide or isopentylamide as P1 residue suggested a covalent binding mode of the boroleucine-derived inhibitors. This was finally proven by crystal structure determination of the most potent inhibitor from this series in complex with the ZIKV protease.
Keywords: NS2B-NS3 protease; Zika virus; boroleucine; crystal structure determination; drug design.
© 2022 The Authors. ChemMedChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
SAR evolution towards potent C-terminal carboxamide peptide inhibitors of Zika virus NS2B-NS3 protease.Bioorg Med Chem. 2022 Mar 1;57:116631. doi: 10.1016/j.bmc.2022.116631. Epub 2022 Jan 23. Bioorg Med Chem. 2022. PMID: 35123179
-
Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.J Med Chem. 2022 May 12;65(9):6555-6572. doi: 10.1021/acs.jmedchem.1c01860. Epub 2022 Apr 27. J Med Chem. 2022. PMID: 35475620
-
Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.Arch Pharm (Weinheim). 2024 Sep;357(9):e2400250. doi: 10.1002/ardp.202400250. Epub 2024 May 29. Arch Pharm (Weinheim). 2024. PMID: 38809037
-
NS2B-NS3 protease inhibitors as promising compounds in the development of antivirals against Zika virus: A systematic review.J Med Virol. 2022 Feb;94(2):442-453. doi: 10.1002/jmv.27386. Epub 2021 Oct 20. J Med Virol. 2022. PMID: 34636434
-
The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection.Int J Mol Sci. 2024 Apr 16;25(8):4376. doi: 10.3390/ijms25084376. Int J Mol Sci. 2024. PMID: 38673962 Free PMC article. Review.
Cited by
-
Recent advances in the study of zika virus structure, drug targets, and inhibitors.Front Pharmacol. 2024 Jul 1;15:1418516. doi: 10.3389/fphar.2024.1418516. eCollection 2024. Front Pharmacol. 2024. PMID: 39011504 Free PMC article. Review.
References
-
- Kraemer M. U. G., R. C. Reiner, Jr. , Brady O. J., Messina J. P., Gilbert M., Pigott D. M., Yi D., Johnson K., Earl L., Marczak L. B., Shirude S., Davis Weaver N., Bisanzio D., Perkins T. A., Lai S., Lu X., Jones P., Coelho G. E., Carvalho R. G., Van Bortel W., Marsboom C., Hendrickx G., Schaffner F., Moore C. G., Nax H. H., Bengtsson L., Wetter E., Tatem A. J., Brownstein J. S., Smith D. L., Lambrechts L., Cauchemez S., Linard C., Faria N. R., Pybus O. G., Scott T. W., Liu Q., Yu H., Wint G. R. W., Hay S. I., Golding N., Nat. Microbiol. 2019, 4, 854–863. - PMC - PubMed
-
- Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V., Vizjak A., Pižem J., Petrovec M., Avšič Županc T., N. Engl. J. Med. 2016, 374, 951–958. - PubMed
-
- Abrams R. P. M., Yasgar A., Teramoto T., Lee M. H., Dorjsuren D., Eastman R. T., Malik N., Zakharov A. V., Li W., Bachani M., Brimacombe K., Steiner J. P., Hall M. D., Balasubramanian A., Jadhav A., Padmanabhan R., Simeonov A., Nath A., Proc. Natl. Acad. Sci. USA 2020, 117, 31365–31375. - PMC - PubMed
-
- Voss S., Nitsche C., Bioorg. Med. Chem. Lett. 2020, 30, 126965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

