DyScore: A Boosting Scoring Method with Dynamic Properties for Identifying True Binders and Nonbinders in Structure-Based Drug Discovery
- PMID: 36327102
- PMCID: PMC9983328
- DOI: 10.1021/acs.jcim.2c00926
DyScore: A Boosting Scoring Method with Dynamic Properties for Identifying True Binders and Nonbinders in Structure-Based Drug Discovery
Abstract
The accurate prediction of protein-ligand binding affinity is critical for the success of computer-aided drug discovery. However, the accuracy of current scoring functions is usually unsatisfactory due to their rough approximation or sometimes even omittance of many factors involved in protein-ligand binding. For instance, the intrinsic dynamics of the protein-ligand binding state is usually disregarded in scoring function because these rapid binding affinity prediction approaches are only based on a representative complex structure of the protein and ligand in the binding state. That is, the dynamic protein-ligand binding complex ensembles are simplified as a static snapshot in calculation. In this study, two novel features were proposed for characterizing the dynamic properties of protein-ligand binding based on the static structure of the complex, which is expected to be a valuable complement to the current scoring functions. The two features demonstrate the geometry-shape matching between a protein and a ligand as well as the dynamic stability of protein-ligand binding. We further combined these two novel features with several classical scoring functions to develop a binary classification model called DyScore that uses the Extreme Gradient Boosting algorithm to classify compound poses as binders or non-binders. We have found that DyScore achieves state-of-the-art performance in distinguishing active and decoy ligands on both enhanced DUD data set and external test sets with both proposed novel features showing significant contributions to the improved performance. Especially, DyScore exhibits superior performance on early recognition, a crucial requirement for success in virtual screening and de novo drug design. The standalone version of DyScore and Dyscore-MF are freely available to all at: https://github.com/YanjunLi-CS/dyscore.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
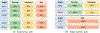

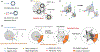


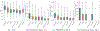
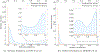






Similar articles
-
Cheminformatics meets molecular mechanics: a combined application of knowledge-based pose scoring and physical force field-based hit scoring functions improves the accuracy of structure-based virtual screening.J Chem Inf Model. 2012 Jan 23;52(1):16-28. doi: 10.1021/ci2002507. Epub 2011 Dec 14. J Chem Inf Model. 2012. PMID: 22017385 Free PMC article.
-
Discrete molecular dynamics distinguishes nativelike binding poses from decoys in difficult targets.Biophys J. 2012 Jan 4;102(1):144-51. doi: 10.1016/j.bpj.2011.11.4008. Epub 2012 Jan 3. Biophys J. 2012. PMID: 22225808 Free PMC article.
-
Task-Specific Scoring Functions for Predicting Ligand Binding Poses and Affinity and for Screening Enrichment.J Chem Inf Model. 2018 Jan 22;58(1):119-133. doi: 10.1021/acs.jcim.7b00309. Epub 2017 Dec 20. J Chem Inf Model. 2018. PMID: 29190087
-
Learning from Docked Ligands: Ligand-Based Features Rescue Structure-Based Scoring Functions When Trained on Docked Poses.J Chem Inf Model. 2022 Nov 28;62(22):5329-5341. doi: 10.1021/acs.jcim.1c00096. Epub 2021 Sep 1. J Chem Inf Model. 2022. PMID: 34469150 Review.
-
Delta Machine Learning to Improve Scoring-Ranking-Screening Performances of Protein-Ligand Scoring Functions.J Chem Inf Model. 2022 Jun 13;62(11):2696-2712. doi: 10.1021/acs.jcim.2c00485. Epub 2022 May 17. J Chem Inf Model. 2022. PMID: 35579568 Free PMC article. Review.
Cited by
-
Protein language models are performant in structure-free virtual screening.Brief Bioinform. 2024 Sep 23;25(6):bbae480. doi: 10.1093/bib/bbae480. Brief Bioinform. 2024. PMID: 39327890 Free PMC article.
-
Morphological profiling for drug discovery in the era of deep learning.Brief Bioinform. 2024 May 23;25(4):bbae284. doi: 10.1093/bib/bbae284. Brief Bioinform. 2024. PMID: 38886164 Free PMC article. Review.
-
BatmanNet: bi-branch masked graph transformer autoencoder for molecular representation.Brief Bioinform. 2023 Nov 22;25(1):bbad400. doi: 10.1093/bib/bbad400. Brief Bioinform. 2023. PMID: 38033291 Free PMC article.
-
miDruglikeness: Subdivisional Drug-Likeness Prediction Models Using Active Ensemble Learning Strategies.Biomolecules. 2022 Dec 23;13(1):29. doi: 10.3390/biom13010029. Biomolecules. 2022. PMID: 36671415 Free PMC article.
-
Recent Advances in Computer-Aided Structure-Based Drug Design on Ion Channels.Int J Mol Sci. 2023 May 25;24(11):9226. doi: 10.3390/ijms24119226. Int J Mol Sci. 2023. PMID: 37298178 Free PMC article. Review.
References
-
- Bhm H-J The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput.-Aided Mol. Des. 1994, 8, 243–256. - PubMed
-
- Feher M Consensus scoring for protein–ligand interactions. Drug DiscovToday 2006, 11, 421–428. - PubMed
-
- Gohlke H; Hendlich M; Klebe G Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000, 295, 337–356. - PubMed
-
- Gohlke H; Klebe G Statistical potentials and scoring functions applied to protein–ligand binding. Curr. Opin. Struct. Biol. 2001, 11, 231–235. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

