A noncanonical RNA-binding domain of the fragile X protein, FMRP, elicits translational repression independent of mRNA G-quadruplexes
- PMID: 36328245
- PMCID: PMC9712993
- DOI: 10.1016/j.jbc.2022.102660
A noncanonical RNA-binding domain of the fragile X protein, FMRP, elicits translational repression independent of mRNA G-quadruplexes
Abstract
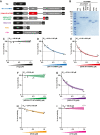
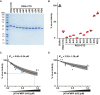
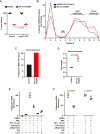
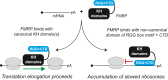
Loss of functional fragile X mental retardation protein (FMRP) causes fragile X syndrome, the leading form of inherited intellectual disability and the most common monogenic cause of autism spectrum disorders. FMRP is an RNA-binding protein that controls neuronal mRNA localization and translation. FMRP is thought to inhibit translation elongation after being recruited to target transcripts via binding RNA G-quadruplexes (G4s) within the coding sequence. Here, we directly test this model and report that FMRP inhibits translation independent of mRNA G4s. Furthermore, we found that the RGG box motif together with its natural C-terminal domain forms a noncanonical RNA-binding domain (ncRBD) that is essential for translational repression. The ncRBD elicits broad RNA-binding ability and binds to multiple reporter mRNAs and all four homopolymeric RNAs. Serial deletion analysis of the ncRBD identified that the regions required for mRNA binding and translational repression overlap but are not identical. Consistent with FMRP stalling elongating ribosomes and causing the accumulation of slowed 80S ribosomes, transcripts bound by FMRP via the ncRBD cosediment with heavier polysomes and were present in puromycin-resistant ribosome complexes. Together, this work identifies a ncRBD and translational repression domain that shifts our understanding of how FMRP inhibits translation independent of mRNA G4s.
Keywords: RNA-binding protein; mRNA; protein synthesis; ribosome; translation control.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. - PubMed
-
- Oberle I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. - PubMed
-
- Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- Yu S., Pritchard M., Kremer E., Lynch M., Nancarrow J., Baker E., et al. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. - PubMed
-
- Niu M., Han Y., Dy A.B.C., Du J., Jin H., Qin J., et al. Autism symptoms in fragile X syndrome. J. Child Neurol. 2017;32:903–909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

