Single-cell RNA sequencing analysis reveals the relationship of bone marrow and osteopenia in STZ-induced type 1 diabetic mice
- PMID: 36328744
- PMCID: PMC9637485
- DOI: 10.1016/j.jare.2022.01.006
Single-cell RNA sequencing analysis reveals the relationship of bone marrow and osteopenia in STZ-induced type 1 diabetic mice
Abstract
Introduction: Type 1 diabetes (T1D) is a multifactorial autoimmune disease. Broad knowledge about the genetics, epidemiology and clinical management of T1D has been achieved, but understandings about the cell varieties in the bone marrow during T1D remain limited.
Objectives: We aimed to present a profile of the bone marrow cells and reveal the relationship of bone marrow and osteopenia in streptozotocin (STZ)-induced T1D mice.
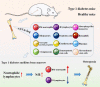
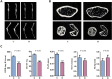
Methods: The whole bone marrow cells from the femurs and tibias of healthy (group C) and STZ-induced T1D mice (group D) were collected for single-cell RNA sequencing analysis. Single-cell flow cytometry and immunohistochemistry were performed to confirm the proportional changes among bone marrow neutrophils (BM-neutrophils) (Cxcr2+, Ly6g+) and B lymphocytes (Cd19+). X-ray and micro-CT were performed to detect bone mineral density. The correlation between the ratio of BM-neutrophils/B lymphocytes and osteopenia in STZ-induced T1D mice was analyzed by nonparametric Spearman correlation analysis.
Results: The bone marrow cells in groups C and D were divided into 12 clusters, and 249 differentially expressed genes were found. The diversity of CD45+ immune cells between groups C and D were greatly affected: the proportion of BM-neutrophils showed a significant increase while the proportion of B lymphocytes in group D showed a significant decrease. X-ray and micro-CT analyses confirmed that osteopenia occurred in group D mice. In addition, the results of single-cell flow cytometry and correlation analysis showed that the ratio of BM-neutrophils/B lymphocytes negatively correlated with osteopenia in STZ-induced T1D mice.
Conclusion: A single-cell RNA sequencing analysis revealed the profile and heterogeneity of bone marrow immune cells in STZ-induced T1D mice for the first time. The ratio of BM-neutrophils/B lymphocytes negatively correlated with osteopenia in STZ-induced T1D mice, which may enhance understanding for treating T1D and preventing T1D-induced osteopenia.
Keywords: Bone marrow; Lymphocyte; Neutrophil; Osteopenia; Single-cell RNA sequencing; Type 1 diabetes.
Copyright © 2022. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Ayhan H., Kasapkara H.A., Aslan A.N., Durmaz T., Keleş T., Akçay M., et al. Relationship of neutrophil-to-lymphocyte ratio with aortic stiffness in type 1 diabetes mellitus. Can J Diabetes. 2015;39(4):317–321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

