Immunomodulatory fecal metabolites are associated with mortality in COVID-19 patients with respiratory failure
- PMID: 36329015
- PMCID: PMC9633022
- DOI: 10.1038/s41467-022-34260-2
Immunomodulatory fecal metabolites are associated with mortality in COVID-19 patients with respiratory failure
Abstract
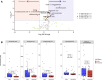
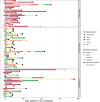
Respiratory failure and mortality from COVID-19 result from virus- and inflammation-induced lung tissue damage. The intestinal microbiome and associated metabolites are implicated in immune responses to respiratory viral infections, however their impact on progression of severe COVID-19 remains unclear. We prospectively enrolled 71 patients with COVID-19 associated critical illness, collected fecal specimens within 3 days of medical intensive care unit admission, defined microbiome compositions by shotgun metagenomic sequencing, and quantified microbiota-derived metabolites (NCT #04552834). Of the 71 patients, 39 survived and 32 died. Mortality was associated with increased representation of Proteobacteria in the fecal microbiota and decreased concentrations of fecal secondary bile acids and desaminotyrosine (DAT). A microbiome metabolic profile (MMP) that accounts for fecal secondary bile acids and desaminotyrosine concentrations was independently associated with progression of respiratory failure leading to mechanical ventilation. Our findings demonstrate that fecal microbiota composition and microbiota-derived metabolite concentrations can predict the trajectory of respiratory function and death in patients with severe SARS-Cov-2 infection and suggest that the gut-lung axis plays an important role in the recovery from COVID-19.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- World Health Organization COVID-19 Dashboard. https://covid19.who.int/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

