GATA-3 is a proto-oncogene in T-cell lymphoproliferative neoplasms
- PMID: 36329027
- PMCID: PMC9633835
- DOI: 10.1038/s41408-022-00745-y
GATA-3 is a proto-oncogene in T-cell lymphoproliferative neoplasms
Abstract
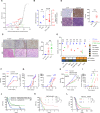
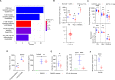
Neoplasms originating from thymic T-cell progenitors and post-thymic mature T-cell subsets account for a minority of lymphoproliferative neoplasms. These T-cell derived neoplasms, while molecularly and genetically heterogeneous, exploit transcription factors and signaling pathways that are critically important in normal T-cell biology, including those implicated in antigen-, costimulatory-, and cytokine-receptor signaling. The transcription factor GATA-3 regulates the growth and proliferation of both immature and mature T cells and has recently been implicated in T-cell neoplasms, including the most common mature T-cell lymphoma observed in much of the Western world. Here we show that GATA-3 is a proto-oncogene across the spectrum of T-cell neoplasms, including those derived from T-cell progenitors and their mature progeny, and further define the transcriptional programs that are GATA-3 dependent, which include therapeutically targetable gene products. The discovery that p300-dependent acetylation regulates GATA-3 mediated transcription by attenuating DNA binding has novel therapeutic implications. As most patients afflicted with GATA-3 driven T-cell neoplasms will succumb to their disease within a few years of diagnosis, these findings suggest opportunities to improve outcomes for these patients.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

