Spatially resolved phosphoproteomics reveals fibroblast growth factor receptor recycling-driven regulation of autophagy and survival
- PMID: 36329028
- PMCID: PMC9633600
- DOI: 10.1038/s41467-022-34298-2
Spatially resolved phosphoproteomics reveals fibroblast growth factor receptor recycling-driven regulation of autophagy and survival
Abstract
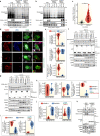
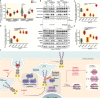
Receptor Tyrosine Kinase (RTK) endocytosis-dependent signalling drives cell proliferation and motility during development and adult homeostasis, but is dysregulated in diseases, including cancer. The recruitment of RTK signalling partners during endocytosis, specifically during recycling to the plasma membrane, is still unknown. Focusing on Fibroblast Growth Factor Receptor 2b (FGFR2b) recycling, we reveal FGFR signalling partners proximal to recycling endosomes by developing a Spatially Resolved Phosphoproteomics (SRP) approach based on APEX2-driven biotinylation followed by phosphorylated peptides enrichment. Combining this with traditional phosphoproteomics, bioinformatics, and targeted assays, we uncover that FGFR2b stimulated by its recycling ligand FGF10 activates mTOR-dependent signalling and ULK1 at the recycling endosomes, leading to autophagy suppression and cell survival. This adds to the growing importance of RTK recycling in orchestrating cell fate and suggests a therapeutically targetable vulnerability in ligand-responsive cancer cells. Integrating SRP with other systems biology approaches provides a powerful tool to spatially resolve cellular signalling.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Cullen PJ, Steinberg F. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018;19:679–696. - PubMed
-
- Hsu VW, Bai M, Li J. Getting active: protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 2012;13:323–328. - PubMed
-
- Sigismund S, Lanzetti L, Scita G, Di Fiore PP. Endocytosis in the context-dependent regulation of individual and collective cell properties. Nat. Rev. Mol. Cell Biol. 2021;22:625–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

